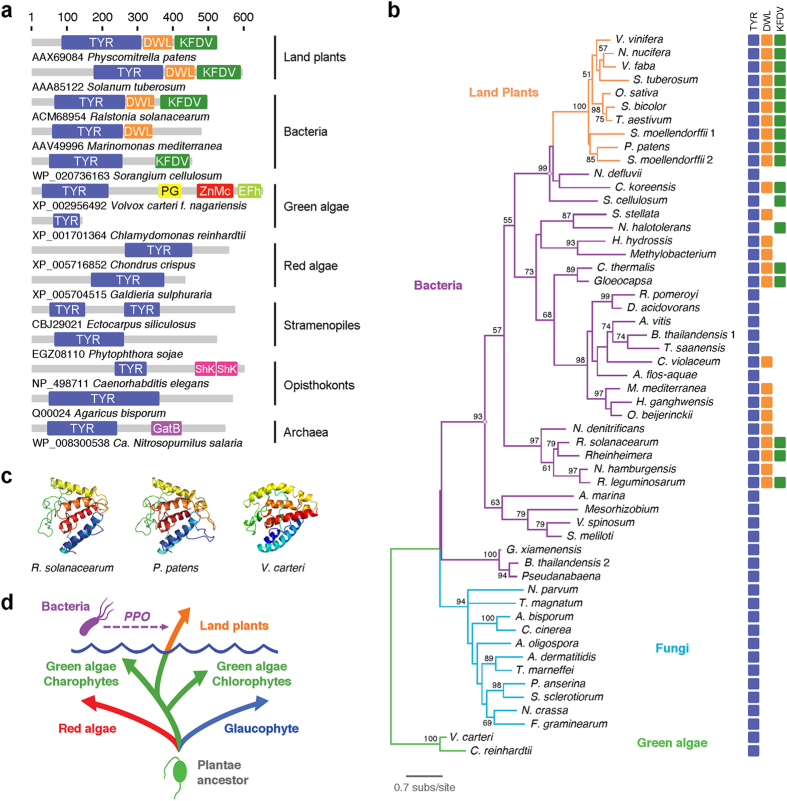
Figure 1. Evidence for horizontal gene transfer of PPO from bacteria to land plants.
(a) Schematic protein domain architecture alignment of different representative proteins that share the tyrosinase domain (TYR; PFAM00264). The characteristic DWL (PFAM12142) and KFDV (PFAM12143) domains of land-plant PPO enzymes are also present with the same architecture in bacterial PPO proteins but appear to be absent in green and red algae, glaucophyta and other eukaryotes. Numbers on top indicate protein (amino acids) length. PG: peptidoglycan-binding domain (PFAM01471). ZnMc: zinc-dependent metalloprotease domain (CD00203). EFh: calcium-binding domain helix-turn-helix (CD00051). ShK: ShK domain-like (PFAM 01549). GatB: GatB domain (PFAM 02637). The predicted conserved domain architecture is illustrated according to CDD results. (b) TYR domain-derived maximum likelihood inference of phylogeny showing that land plant PPO-TYR domains form a well-supported monophyletic group with several bacteria. Taxa and branches are colour coded with fungi in light blue, bacteria in purple, land plants in orange and green algae in green. Circles indicate well-supported nodes of the plant-containing bacterial branch. Bootstrap support values above 50 are shown. Domains found in the proteins of the tree are shown as boxes on the right. (c) Structures of TYR domains of R. solanacearum, P. patens and V. carteri. Models are independently displayed based on rainbow colouring scheme (N-terminal coloured blue and C-terminal coloured red). The analysis was performed with 100% of residues modelled at >90% confidence. (d) Scheme of the PPO bacteria-to-land plants horizontal gene transfer hypothesis.