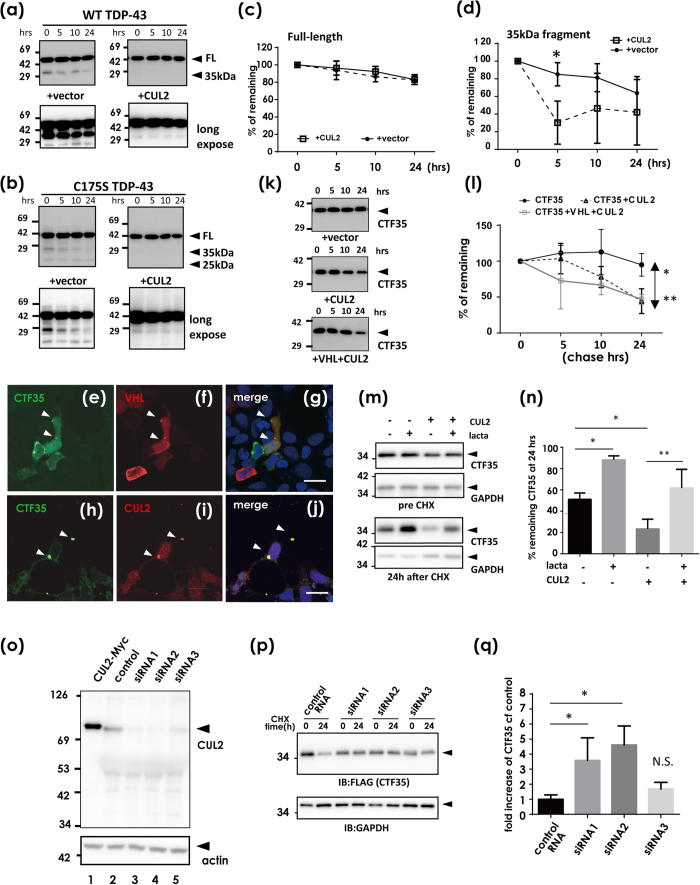
Figure 4. Fragmentation-mediated clearance of TDP-43 by CUL2 E3 ligase.
(a–d) Protein degradation assay of WT TDP-43 (a,c,d) and C175S mutant (b) with or without CUL2. At 48 h after transfection of TDP-43-FLAG with or without Myc-CUL2, cells were treated with 100 μg/ml cycloheximide (CHX). The samples were harvested 0, 5, 10, and 24 h after CHX treatment. (a) Western blot using anti-FLAG antibodies. (c,d) A densitometry for FL (c) and 35-kDa (d) WT TDP-43. Differences were evaluated by two-way ANOVA (mean ± SD from three independent experiments; *p < 0.05). (e–n) CUL2 promotes CTF35 degradation at proteasomes. (e–j) Confocal laser micrographs showing the colocalization of CTF35 and VHL (e,g) or CUL2 (h–j). CTF35-FLAG together with HA-VHL or Myc-CUL2 was transiently co-transfected into HEK293A cells and analyzed by double immunofluorescence. Scale bar, 20 μm. Arrowheads show colocalized inclusions. (k) Western blotting for chase study of CTF35 using anti-FLAG antibodies, and densitometry (l). Differences were evaluated by two-way ANOVA (mean ± SD from three independent cultures; *p < 0.05, **p < 0.01). (m,n) The effect of proteasome inhibition on CUL2-mediated CTF35 clearance. At 48 h after transfection of CTF35-FLAG with or without Myc-CUL2, cells were cultured in the presence or absence of lactacystin (10 μM, 8 h). (m) Western blot. (n) The average percentage of reduction from the amount of time zero. Differences were evaluated by one-way ANOVA (mean ± SD from three independent cultures; *p < 0.05, **p < 0.01). (o–q) Effect of CUL2 knockdown on the amount of CTF35. HEK293A cells were treated with three siRNAs or control oligo for CUL2 for 48 h. (o) Western blot using anti-CUL2 antibody. (p,q) The chase assay of CTF35 during CUL2 knockdown. (p) A representative Western blot using anti-FLAG and anti-GAPDH antibodies. (q) The increased ratio of CTF35 in CUL2-knockdown cells compared with control oligo. The similar trends were observed in three independent studies. Differences were evaluated by one-way ANOVA (mean ± SD from triplicate experiments; *p < 0.05).