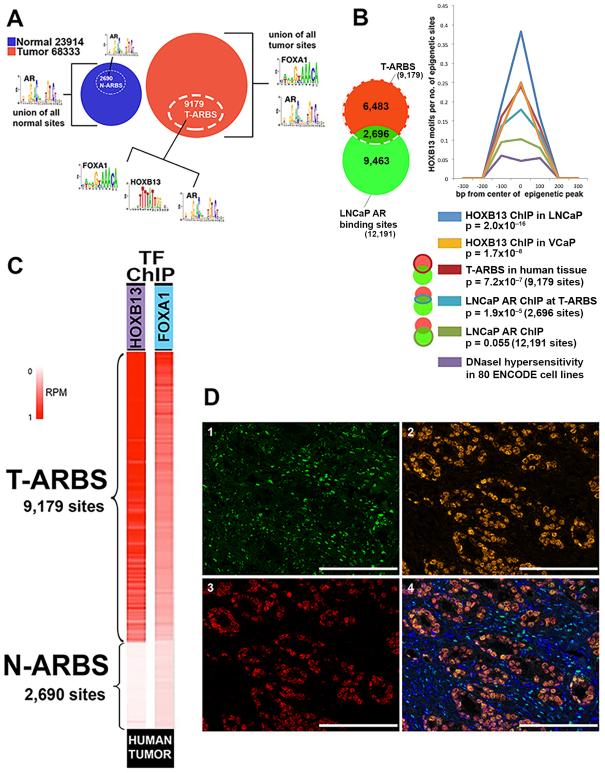
Fig. 3. FOXA1 and HOXB13 co-localize with AR at tumor-specific AR sites.
A. DNA binding motif analysis in subsets of the overall cohort - normal prostate tissue (blue circle), tumor tissue (red circle), and the T-ARBS (white dashed circle). B. At the set of LNCaP AR binding sites that overlap with the set of human T-ARBS, the HOXB13 binding motif emerges as statistically significant. The HOXB13 binding motif did not achieve significance when interrogating the entire LNCaP AR cistrome. At right, enrichment of HOXB13 motif across six conditions relative to background. Background is calculated as HOXB13 motifs in DNaseI hypersensitivity sites derived from 80 cell types included in the ENCODE project. C. HOXB13 and FOXA1 ChIP-seq in human prostate tumor specimens showed that these transcription factors (transcription factors) co-localize with T-ARBS. D. Representative core showing nuclear co-localization of AR, FOXA1 and HOXB13 protein by multi-plex immunohistochemistry staining in prostate cancer tissue: green, AR expression (1); orange, FOXA1 expression (2); red, HOXB13 expression (3); Multispectral image for AR, FoxA1 and HOXB13, with DAPI counterstained for nuclear masking, demonstrating AR, FOXA1 and HOXB13 nuclear co-localization in prostate cancer cells (4). Scale bars represent 100 μM.