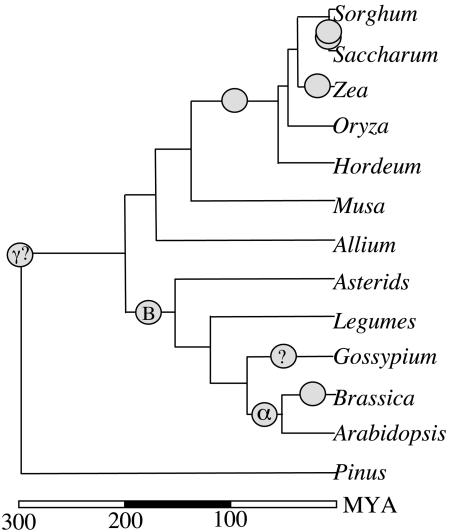
Fig. 3.
An early phylogenetic tree of genomic duplications for the angiosperms. By integrating data described herein and elsewhere (7), ancient duplications in the monocots and dicots, respectively, are superimposed on a partial angiosperm phylogenetic tree that also represents well established recent duplications in several lineages. Open circles indicate possible chromosomal duplication or polyploid formation events. Question marks indicate (i) the need for additional data to support tentative indications of a polyploidization event in the Gossypium lineage (J. Rong, J.E.B., and A.H.P., unpublished data) and (ii) uncertainty about the dating of the γ event (7). Gene tree analyses (see text and Table 1) support largely “one-to-one” correspondence of the rice chromosomes to those of other diploid cereals, but suggest the need for “one-to-two (or more)” comparisons to more distant lineages. More recent duplications and/or polyploid formation within many lineages further complicate comparative genomics. Branch lengths along the y axis approximate divergence times cited (7) or Ks data reported herein, converted to MY (millions of years) by using the average of current molecular clocks (18, 19).