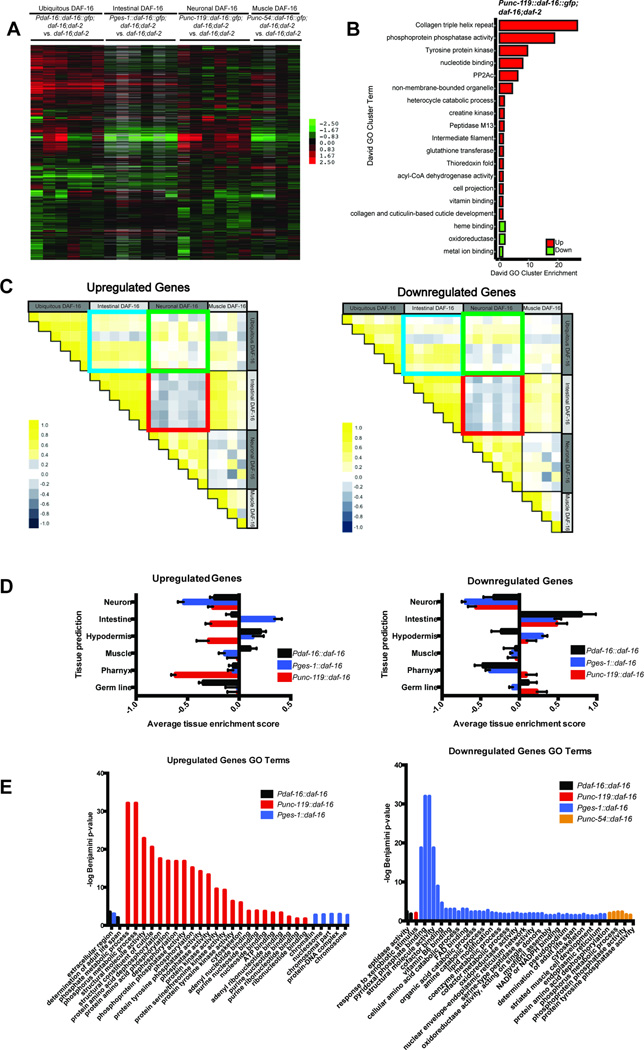
Extended Data Fig 1.
A) DAF-16 tissue-specific transgenics; heat map of all genes with expression differences ≥1.5-fold in ≥3 arrays. B) Significant Gene Ontology (GO) cluster terms from Punc-119::daf-16-regulated up- and down-regulated genes (Enrichment score >1). C) Pairwise Pearson correlations between arrays of DAF-16-up-regulated or down-regulated targets. The red box highlights the negative correlation between neuronal DAF-16 rescued targets (Punc-119::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2) and intestinal DAF-16 targets (Pges-1::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2), while the blue box shows the positive correlation between intestinal DAF-16 targets (Pges-1::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2) and whole worm DAF-16 targets (Pdaf-16::daf-16::gfp; daf-16;daf-2 vs daf-16;daf-2). The green box shows the weak correlation between neuronal rescued and whole worm DAF-16 targets. D) Tissue enrichment analysis (Mean ± SEM) of significant DAF-16-rescued up- and down-regulated genes (Supplementary Table 1) (FDR < 0.5). E) Significant Gene Ontology (GO) terms (adjusted p-value < 0.05) for DAF-16 up-regulated and down-regulated genes from whole worm, intestine-, neuron-, and muscle-rescued DAF-16 strains. Genes used for GO analysis (Supplementary Table 2) were derived from SAM analysis of the microarrays in (A) and Supplementary Table 1.