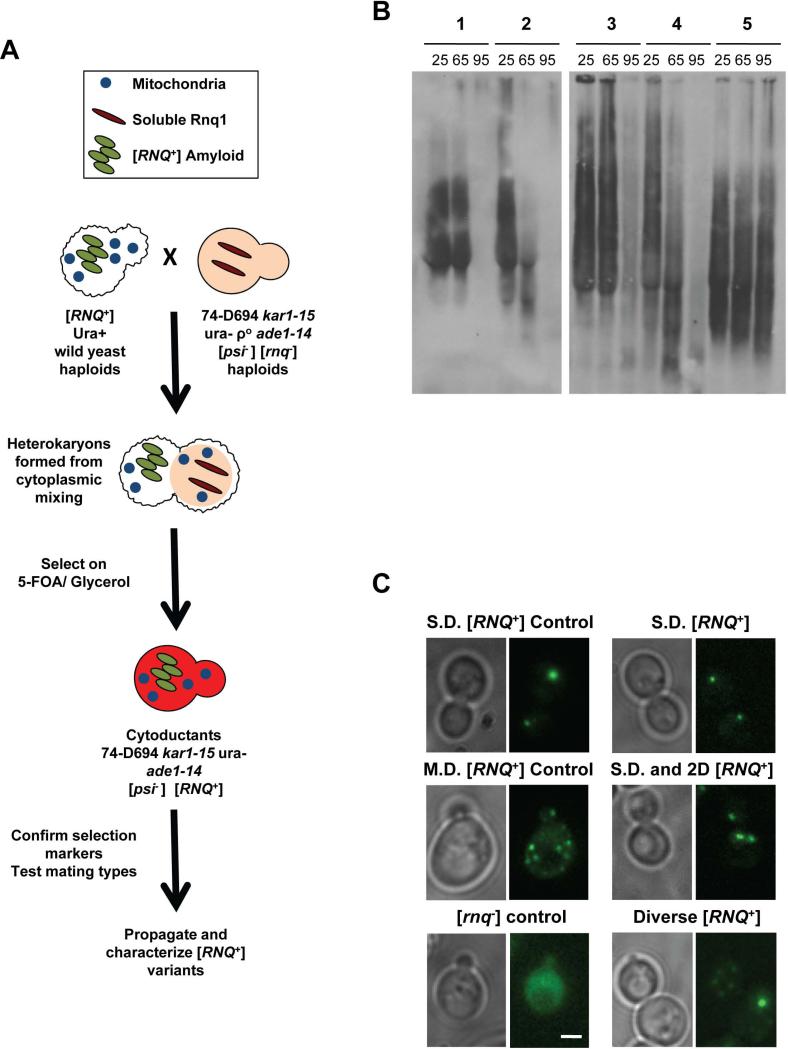
Figure 1. [RNQ+] variants can be distinguished by differences in temperature sensitivity and by cellular aggregate distribution.
A. Schematic depicting cytoplasmic transfer of [RNQ+] from yeast of wild origin into the [psi−] [rnq−] strain 74-D694, as detailed in Materials and Methods. B. Cell lysates from 74-D694 yeast cytoduced with wild [RNQ+] prions were incubated for 7 minutes at 25°C, 65°C or 95°C. Lysates were then subjected to SDD-AGE and western blotting using an anti-Rnq1 antibody. Example yeast strains containing aggregates with different temperature sensitivities are shown and are listed as follows: 1) no breakdown at 65°C, eliminated at 95°C, 2) breakdown at 65°C, eliminated at 95°C, 3) no breakdown at 65°C, remain at 95°C, 4) 74-D694 Multi-Dot High [RNQ+] control: breakdown at 65°C, eliminated at 95°C, 5) 74-D694 Single-Dot High [RNQ+] control: no breakdown at 65°C, remain at 95°C. C. 74-D694 yeast that were cytoduced with wild [RNQ+] prions were transformed with the Rnq-GFP plasmid and imaged as detailed in Materials and Methods. Two 74-D694 cytoductants for each wild [RNQ+] strain were examined in three independent experiments. More than 100 cells were analyzed for each wild strain. Photos are Z stacks of a representative subset of cells. M.D.= multiple fluorescent puncta, 2.D.= two fluorescent puncta, S.D.= single fluorescent puncta. Scale bar in the lower right corner of the [rnq−] control= 2μm and applies to all panels.