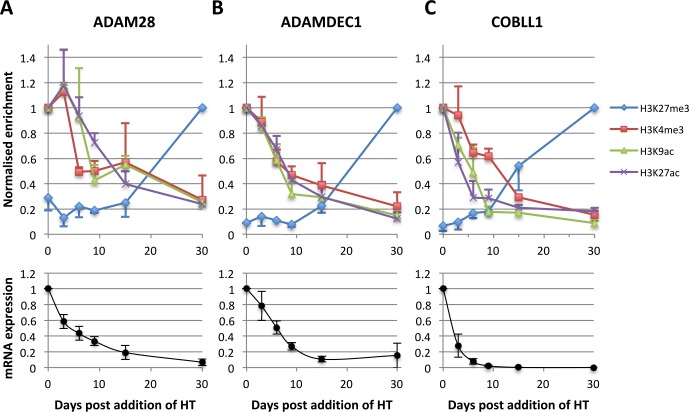
Fig 5. Correlation between changes of epigenetic marks over time at the TSS of ADAM28, ADAMDEC1 and COBLL1 with gene expression.
ChIP-qPCR values (top) at the TSS of ADAM28 (A), ADAMDEC1 (B) and COBLL1 (C) from 3CHT A13 and 3CHT C19 time-courses were normalised to day 30 +HT for the repressive histone mark H3K27me3 and to day three -HT for the activation-associated histone marks H3K4me3, H3K9ac and H3K27ac, which were then set as day 0. Mean values ± standard deviation from both replicate time-courses are shown. For better visual clarity, error bars are colour-matching the corresponding histone mark and only upper bars are shown for activation-associated marks and lower bars for H3K27me3. mRNA expression data for each gene (bottom) are shown as mean values ± standard deviation from three replicate time courses (3CHT A13 rep 1+2 and 3CHT C19).