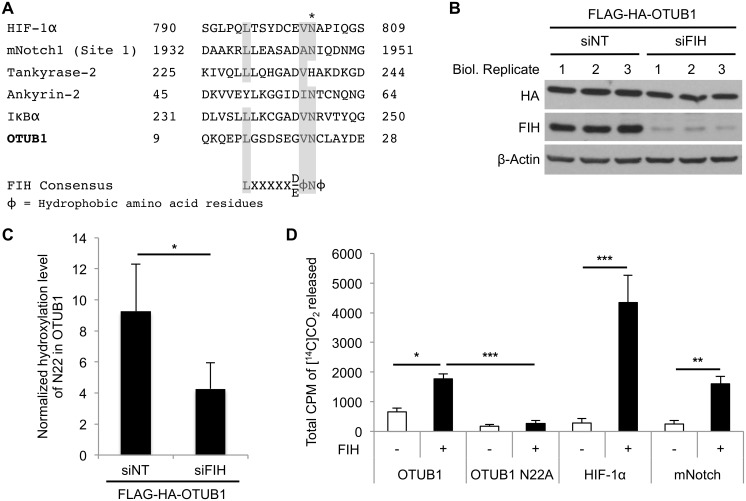
Fig 3. FIH hydroxylates OTUB1 on asparagine 22.
(A) Sequence alignment of OTUB1 with previously identified FIH target proteins reveals that N22 lies within an FIH consensus motif in OTUB1. Amino acid residues that are part of the FIH consensus sequence in human OTUB1 are highlighted in gray. The FIH-targeted N22 residue of human OTUB1 is highlighted with “*”. (B) Western blot analysis demonstrating effective knockdown of FIH in HEK293 cells. (C) Hydroxylation levels of OTUB1 N22 of the samples shown in (B) after FLAG-specific immunoprecipitation. The detected hydroxylation peptide intensity of each sample was normalized to the amount of non-hydroxylated OTUB1 N22 peptide intensity detected. (D) In vitro CO2 capture assay for MBP-hFIH-1-catalyzed hydroxylation-coupled stoichiometric release of 14CO2 from [1-14C]-2-oxoglutarate with synthesized peptides as FIH substrates containing demonstrated hydroxylation sites of HIF-1α, NOTCH and the putative hydroxylation site (N22) of OTUB1. An OTUB1 peptide containing a N22A point mutation was used as control for an N22-specific reaction. The experiment described in (C) was performed with three biological replicates and two technical replicates per sample and data are presented as mean + SD; * p < 0.05 by Student’s t test. Data presented in (D) as mean + SD of n = 3 experiments; * p < 0.05, ** p < 0.01, *** p < 0.001 by one-way ANOVA followed by Tukey post test. The underlying data of panels C and D can be found in S1 Data.