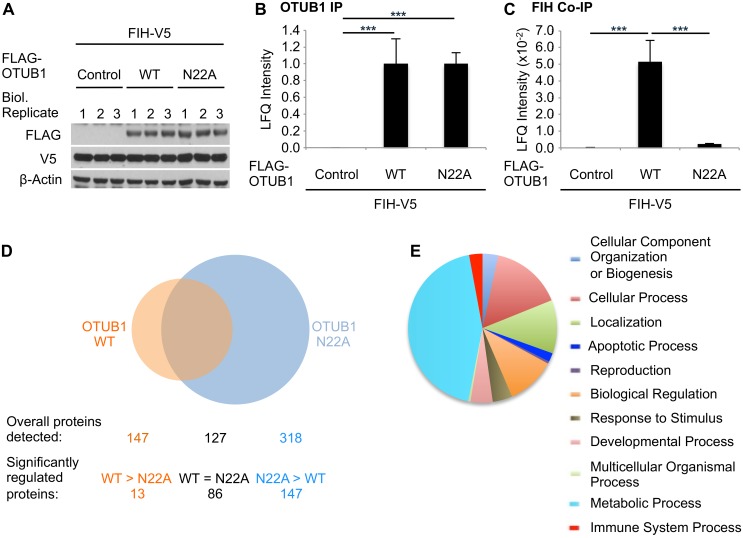
Fig 4. Point mutation of asparagine 22 to alanine regulates the OTUB1 interactome.
HEK293 cells were transfected in biological triplicates with pFIH-V5 and empty vector (control), pFLAG-OTUB1 WT, or pFLAG-OTUB1 N22A for 24 h. Following FLAG immunoprecipitation, the precipitates were analyzed for OTUB1-associated proteins by mass spectrometry. (A) Western blot analysis of overexpression levels of OTUB1 WT and N22A. (B) Relative amount of immunoprecipitated OTUB1 in each sample. (C) Relative amount of FIH co-precipitated with OTUB1 in each sample (normalized to precipitated OTUB1 levels). (D) Number of interacting proteins enriched in FLAG-OTUB1 WT or N22A over control. “Significantly regulated proteins” indicates the number of proteins being significantly different between FLAG-OTUB1 WT and N22A as indicated. Peptide sequences that were assigned to several proteins were counted as a single interaction. (E) Gene Ontology analysis for biological processes of proteins significantly enriched and increased by at least 2-fold in FLAG-OTUB1 N22A over FLAG-OTUB1 WT. The experiment was performed with three biological replicates and two technical replicates per sample. (B), (C) Data presented as mean + SD. *** p < 0.001 by Student’s t test. (D), (E) The analyses were performed using Panther database (www.pantherdb.org).