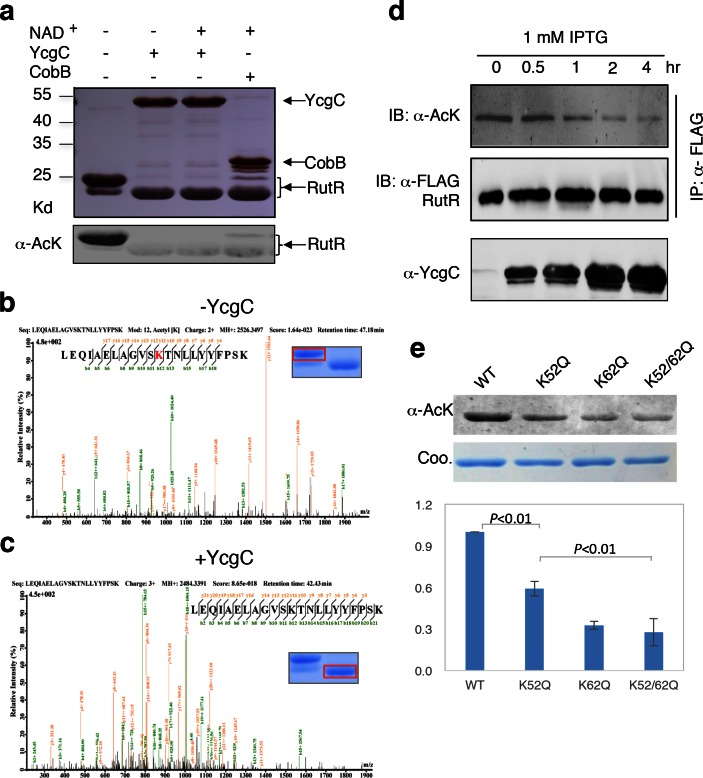
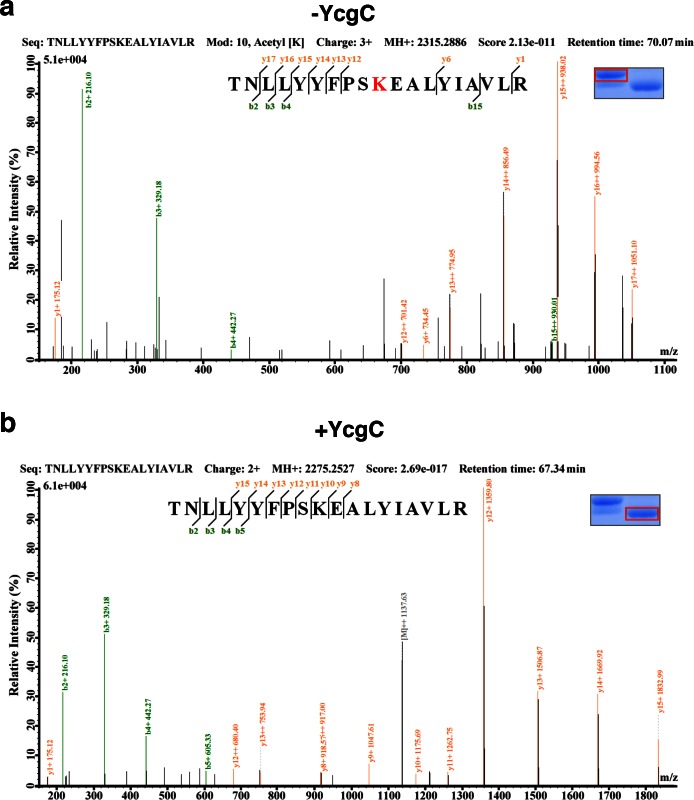
Figure 2. In vitro and in vivo characterization of YcgC’s KDAC activity.
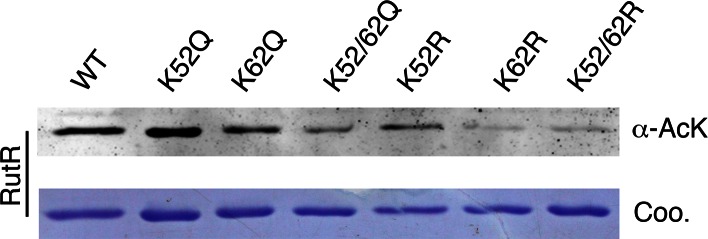
(a) In vitro assays of the KDAC activity of YcgC on RutR demonstrated that its KDAC activity does not require either NAD+ or Zn2+ as cofactors. Incubation with YcgC almost completely abolished the slower migrating acetylated RutR bands (upper panel) as evidenced by immonublotting (lower panel). (b,c) LC-MS/MS analysis to determine the residues of RutR deacetylated by YcgC. RutR was treated with YcgC first and the untreated RutR used as the control. Both these two samples were resolved on a SDS-PAGE gel side by side. The upper band represents the Kac-containing starting materials and the lower band represents the K-containing product, which were then recovered from the gel and subjected for MS/MS analysis (inserts). Lys52 was identified as an acetylated site in RutR protein (b). After incubating with YcgC, acetylation on K52 was no longer detectable (c). (d) RutR is deacetylated by YcgC in Escherichia coli. A 3xFLAG tag was chromosomally inserted at the 3′-end of rutR coding sequence. Acetylation of 3xFLAG-tagged RutR was monitored upon induction of YcgC. While the protein level of RutR was unchanged (middle panel), its acetylation level was dramatically reduced as a function of YcgG induction (upper panel). YcgC’s expression was monitored using a custom-made antibody (lower panel). (e) Mutagenesis of RutR confirmed that K52 and K62 are acetylated in vivo. Two single mutants K52Q and K62Q and one double mutant K52/62Q were constructed. These mutants along with WT RutR were produced and purified in E. coli. Equal amounts of purified proteins were Western blotted with the α-AcK antibody, quantitation of acetylation level of these samples were performed. KDAC: Lysine deacetylase; LC-MS/MS: Liquid chromatography–mass spectrometry; IP: Immunoprecipitation.
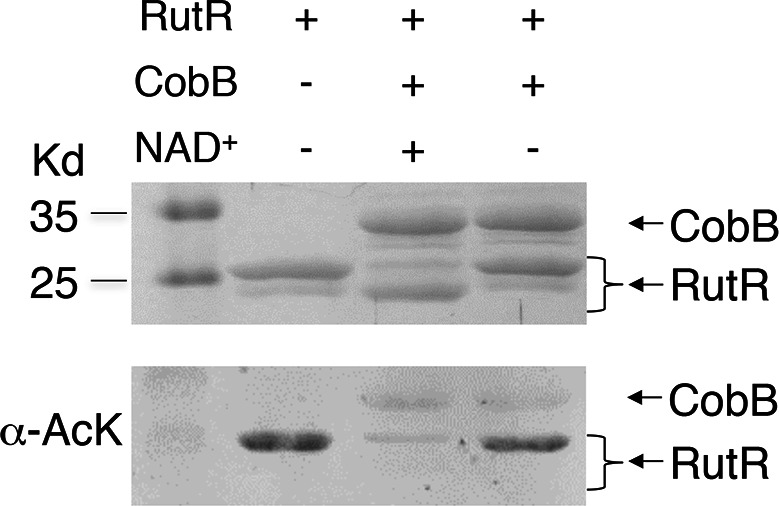
Figure 2—figure supplement 1. Without NAD+ , CobB could not deacetylate RutR.