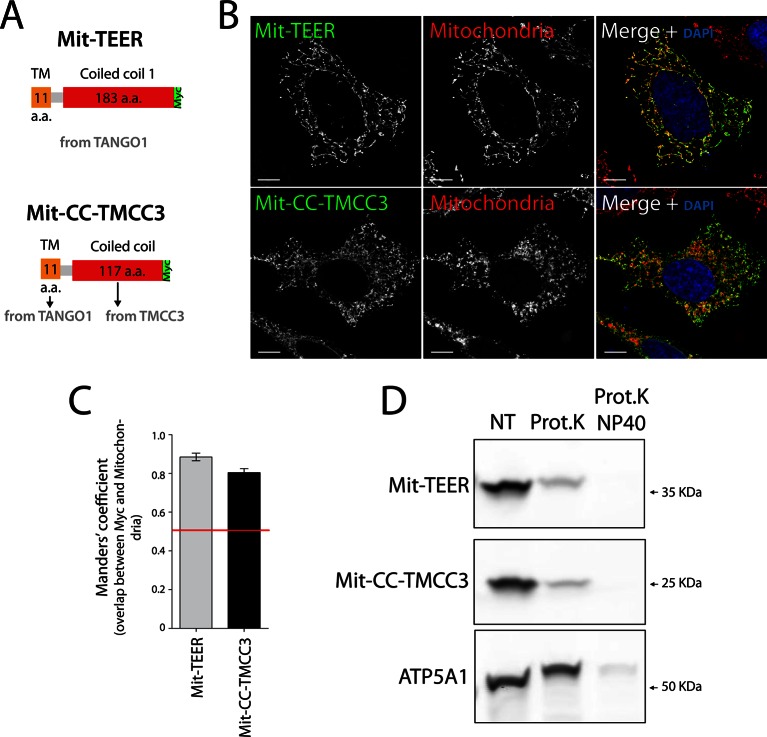
Figure 4. The sequence and topology of mitochondrially targeted TEER (Mit-TEER) and CC-TMCC3 (Mit-CC-TMCC3).
(A) A scheme depicting the coding regions of the two constructs for mitochondrial targeting - Mit-TEER-Myc (residues 1187–1396 of TANGO1, including 11 amino acids of its transmembrane domain, tagged with Myc) or Mit-CC-TMCC3-Myc (the same 11 amino acids of TANGO1’s transmembrane domains and residues 282–398 of TMCC3, tagged with Myc). (B) HeLa cells were seeded on coverslips and transfected with Mit-TEER-Myc or Mit-CC-TMCC3-Myc for 48 hr. To visualize mitochondria (red), cells were either incubated with the Mito-tracker dye for 30 min in serum-free medium before fixation or stained with an anti-ATP5A1 antibody after fixation and permeabilisation. They were also stained with DAPI (blue) and immunostained for Myc (green). (C) Colocalisation quantifications were calculated by Manders’ correlation coefficient by measuring the overlap between the green and the red channels. Error bars reflect the standard error of the mean (SEM) of more than 20 cells from at least three independent experiments. Scale bars correspond to 10 µm. ***P <0.001. (D) Mitochondrial membranes were isolated from HeLa cells transfected with either Mit-TEER or Mit-CC-TMCC3 and split into three fractions. The first was kept as a control (NT); proteinase K (0.1mg/ml) (Prot.K) was added to the second fraction; and to the third fraction, both proteinase K and the detergent NP40 (at a final concentration of 1%) were added (Prot.K/NP40). Samples were incubated on ice for one hour and western blotted with an anti-Myc antibody, to detect Mit-TEER and Mit-CC-TMCC3, and an anti-ATP5A1, a subunit of the ATP synthase, localized to the mitochondrial inner membrane. The loss of Mit-TEER/Mit-TMCC3 but not of ATP5A1, in the absence of detergent, indicated that these constructs are exposed to the cytoplasm.