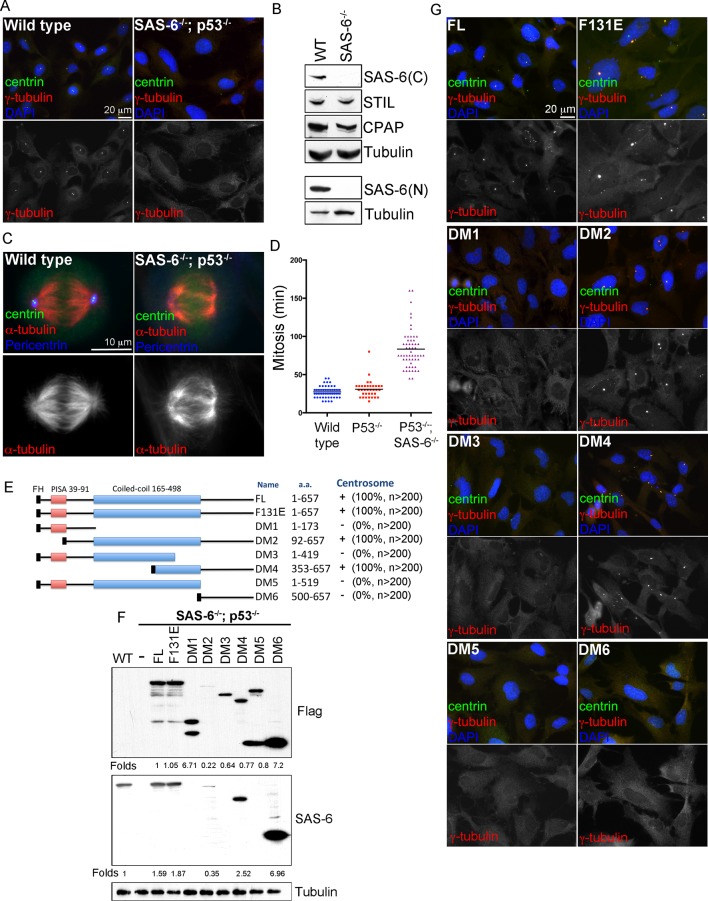
Figure 1. De novo centrosome formation in the absence of SAS-6 self-oligomerization. .
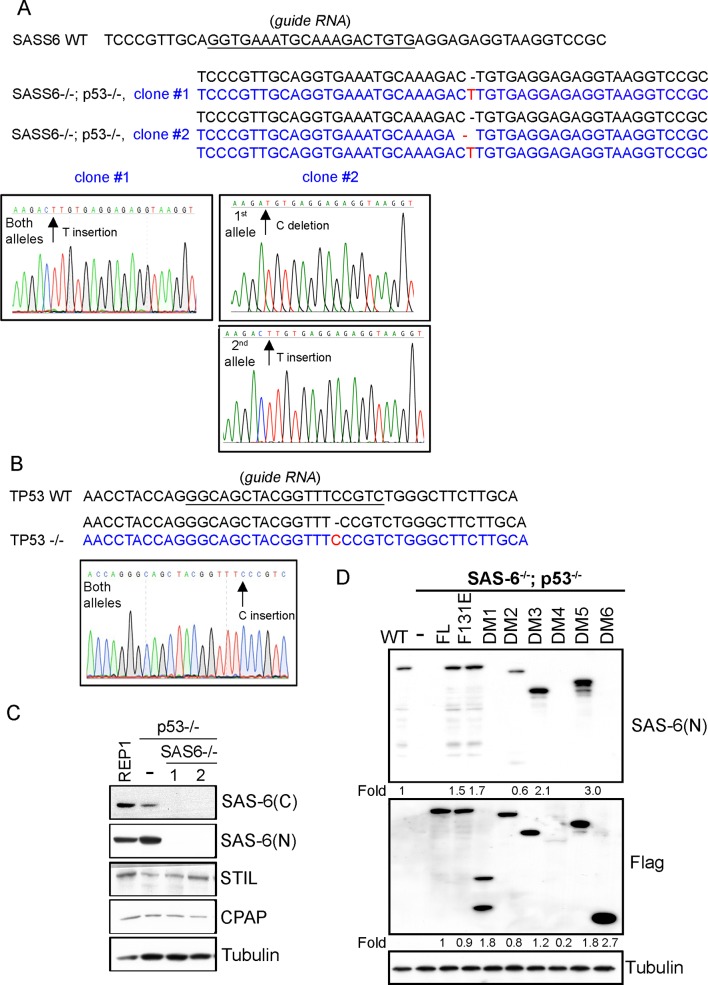
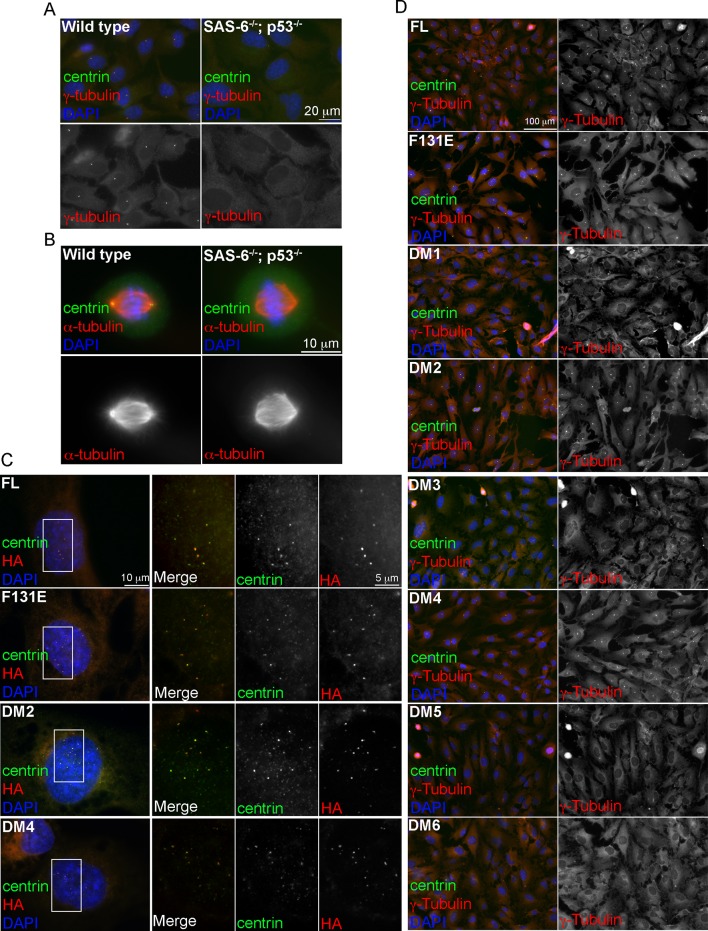
(A) Wild-type (WT) or acentrosomal (p53-/-; SAS-6 -/-; clone #1) RPE1 cells were stained with anti-centrin (green) and γ-tubulin (red). DNA (DAPI, blue). (B) Western blot analysis of WT or SAS-6-/- cells with indicated antibodies, including both N- and C-terminal SAS-6 antibodies (SAS-6N; SAS-6C). (C) WT or acentrosomal cells in mitosis were stained with anti-centrin (green), α-tubulin (red), and pericentrin (blue). (D) The duration of mitosis in WT or SAS-6-/- cells was measured through time-lapse imaging of live cells. (E) A schematic diagram showing various SAS-6 mutants tagged with Flag and HA (FH). Clone #1 p53-/- SAS-6-/- cells were infected with lentiviruses carrying various of SAS-6 constructs, and the ability of each construct in rescuing centrosome formation was indicated and quantified in infected cells expressing detectable HA-tagged SAS-6. n, number of infected cells examined. (F) Isogenic SAS-6-/-; p53-/- cells carrying indicated SAS-6 constructs (see Methods) were induced for SAS-6 expression for 3 days and then analyzed by western blot with Flag antibodies or C-terminal SAS-6 antibodies (SAS-6C). For analyses with N-terminal SAS-6 antibodies (SAS-6N), please see Figure 1—figure supplement 1D. Note that in Flag staining, DM6 leaked to the lane of DM5. The expression level of each SAS-6 construct, indicated as fold changes (Folds) relative to the endogenous SAS-6 or SAS-6FL, is shown. (G) Isogenic SAS-6-/-; p53-/- cells carrying indicated SAS-6 constructs were treated as (F) and analyzed by immunofluorescence microscopy using indicated antibodies (Scale bar: 20 μm in A and G, 10 μm in C).