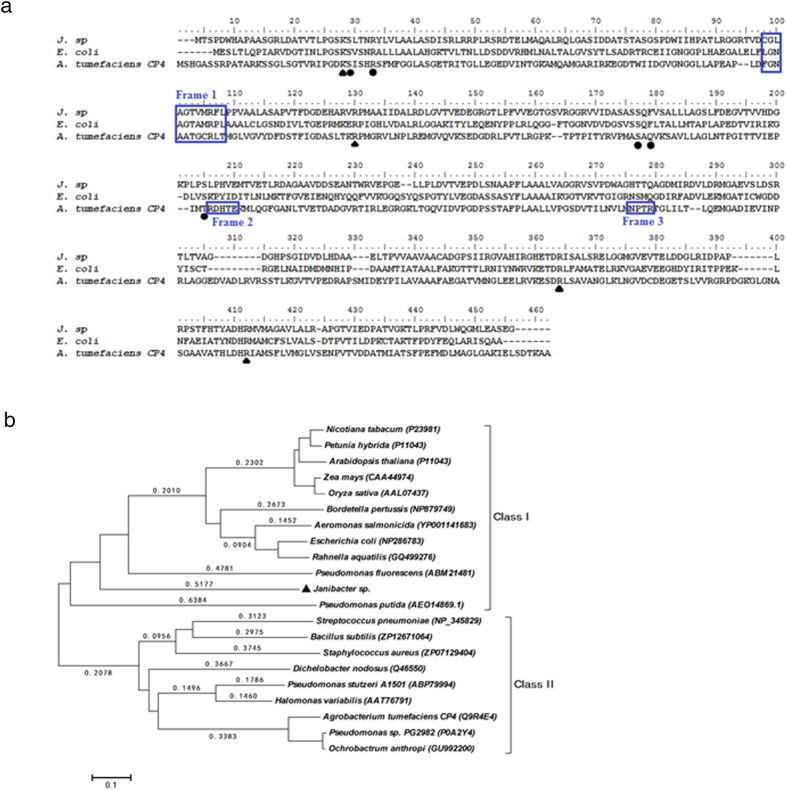
Figure 1.
(a) Multiple alignments of amino acid sequences from aroAJ. sp with representative class I and class II aroA enzymes using ClustalW program. Triangles, residues critical for PEP binding; and circles, residues critical for S3P binding. The two regions involved in glyphosate resistance in class II aroA enzymes are boxed in Frames 2 and 3. The motif important for interaction with PEP conserved in class I aroA enzymes is boxed in Frame 1. (b) The phylogenetic tree was based on homologous sequences of the aroA proteins and the neighbor-joining methods (MEGA4.0). The percentage of the tree from 1000 bootstrap resamples supporting the topology is indicated when above 50. Accession numbers or international patent publication numbers are shown in parentheses. The scale bar represents 0.1 substitutions per position.