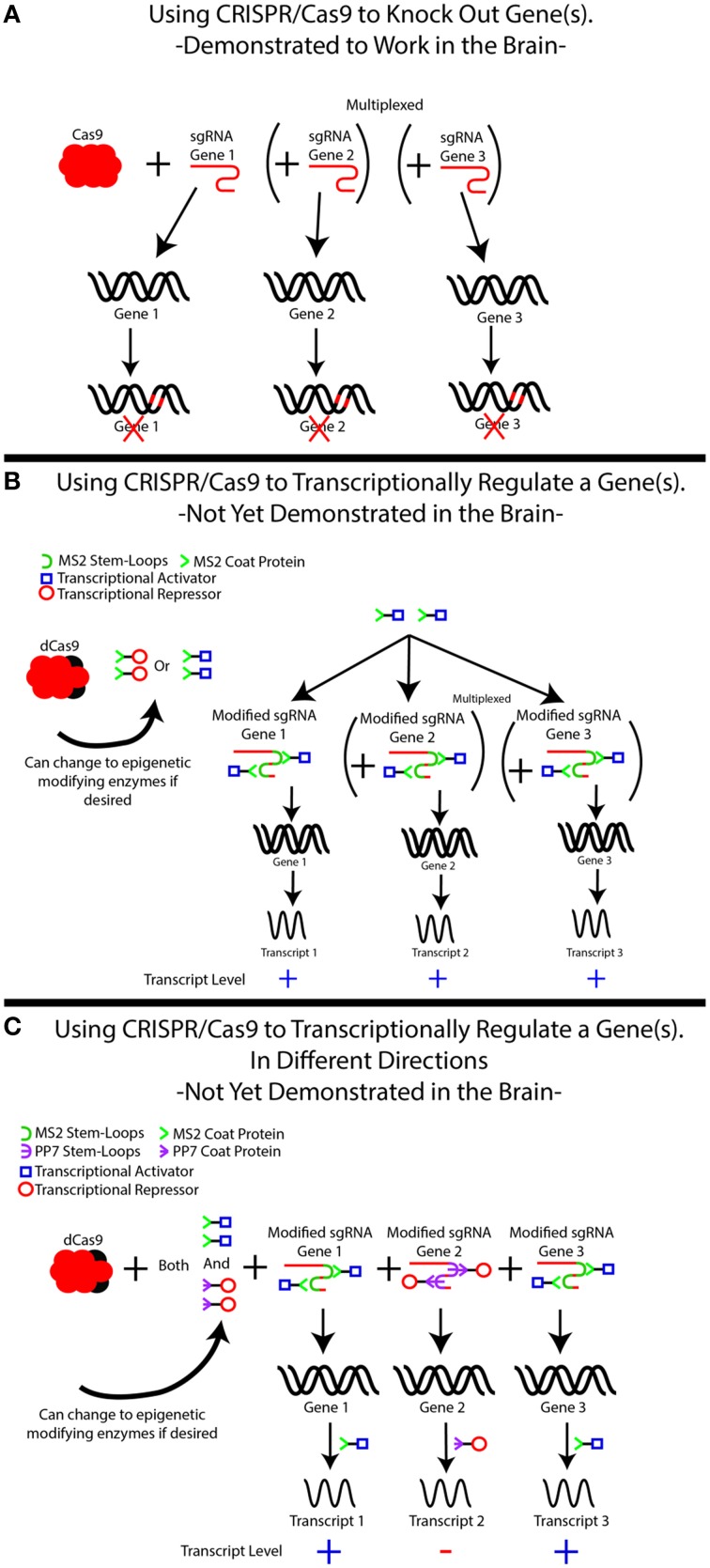
Figure 1.
The use of CRISPR/Cas9 for simplexed and multiplexed gene modulation. (A) CRISPR/Cas9-based gene knockouts have been demonstrated in the brain in both simplexed (1 target) and multiplexed (≥2 genes) fashions. Although our example demonstrates three gene targets, additional targets may be added. In each case, knockdown is achieved by creating a single guide RNA (sgRNA) specific to the gene of interest, which recruits Cas9 to cut the DNA in order to induce frameshift mutations that knock out the gene. Frameshift mutations are illustrated as red marks on the DNA. The use of multiple sgRNAs can induce knockouts of multiple genes simultaneously. (B) An inactive form of Cas9, known as dCas9, can be used to modulate gene expression by recruiting transcriptional regulators to genes of interest, but this system has yet to be demonstrated in the brain. In this system, the sgRNA–dCas9 molecule is treated as a scaffold for recruiting other proteins, most commonly transcriptional activators (blue squares) or transcriptional repressors (red circles) to the specified location within the genome. This is not limited to transcriptional regulators and can easily be modified to bring epigenetic-modifying enzymes or specific transcription factors to these locations. This is accomplished by modifying the sgRNA to include MS2 sequences (green half circle), which is recognized by the MS2 coat protein, MCP (green >). The MS2 coat protein is fused to the protein of interest, in this case either a transcriptional activator or repressor. The MS2-fusion protein is transduced into the cells of interest in addition to the dCas9 and the modified sgRNA, directing the activator/repressor to the specific loci. It is important to note that with only an MS2-fusion construct, only 1 type of fused protein can be delivered. That is, either transcriptional activation or repression can occur, but not both. Simultaneous multiplexing of repressors and activators to distinct loci is demonstrated in (C), where additional stem loops (the PP7 stem loop, purple ϶) and stem loop binding proteins (PP7 binding protein, purple  ) are added to the system and fused to specific DNA-modifying enzymes. This allows separate proteins to be recruited to distinct loci, achieving mixed activation/repression of different genes.
) are added to the system and fused to specific DNA-modifying enzymes. This allows separate proteins to be recruited to distinct loci, achieving mixed activation/repression of different genes.