Figure 3.
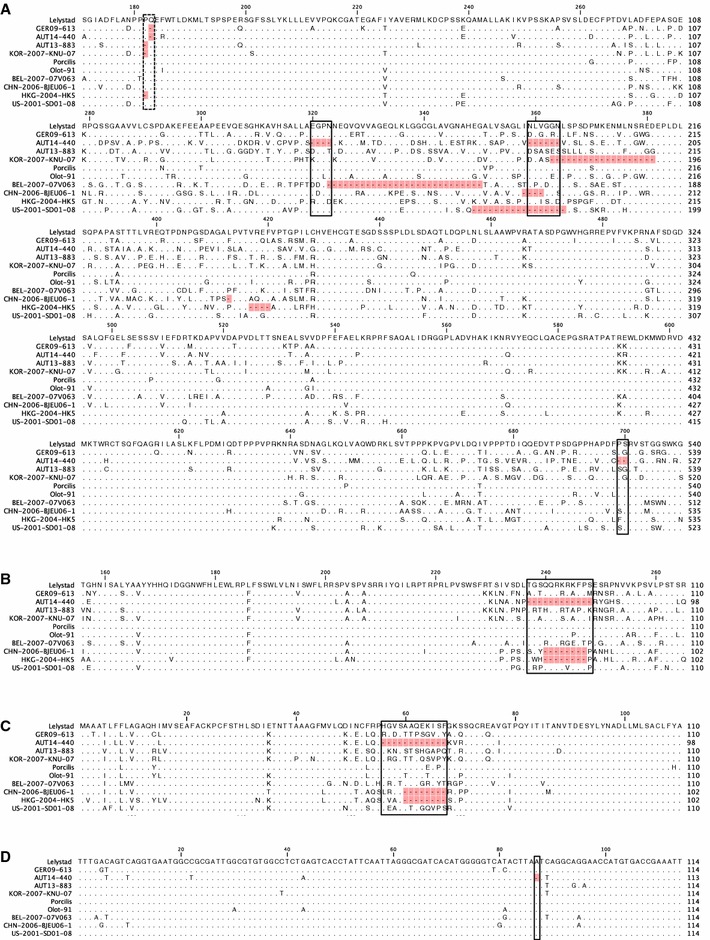
Multiple alignments of partial nsp2 (A), GP3 (B) and GP4 (C) amino-acid sequences of eleven different European PRRSV strains. Only aa differing from Lelystad virus (LV) are shown, with identical aa represented by dots. Deletions compared to LV are marked in red. Deletions of AUT14-440 are highlighted with solid boxes; a deletion in all three isolates is indicated with a dotted box. The numbers above the alignment indicate the position in the protein. A A single aa deletion is present in all three isolates and KNU-07 at position 182 or 183 in nsp2. In AUT14-440 aa 320–323, aa 359–364 and aa 699–700 are deleted compared to LV. B, C The box marks a 12 aa deletion in the overlapping region of GP3 and GP4 of AUT14-440. D Multiple alignment of 3′-UTR nucleotide sequences of ten European PRRSV strains. Nucleotides differing from Lelystad virus (LV) are shown, with identical nucleotides represented by dots. A deletion of 1 nt at position 87 in AUT14-440 is marked in red and highlighted with a solid box.
