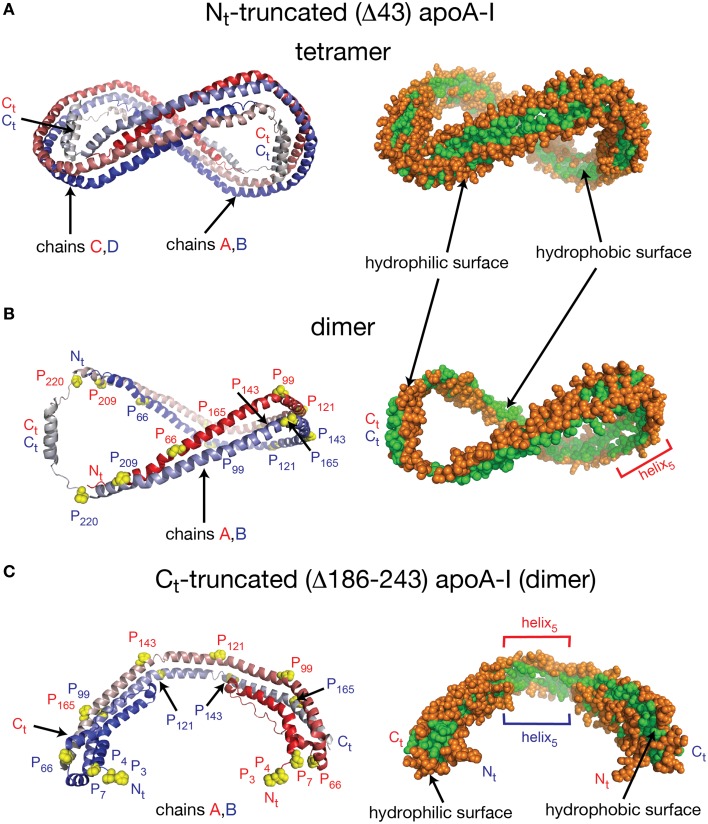
Figure 1.
Crystal structures of truncated lipid free apoA-I. Both crystal structures have the apoA-I chains orientated antiparallel in helix 5 registry, i.e., the helix 5 domains (P121–S142) from both chains are on top of each other. (A) Left: Cartoon representation of the crystal structure of the Δ43–apoA-I (Nt truncated apoA-I) tetramer (PDB id: 1AV1). The structure shows two twisted apoA-I dimers (A,B and C,D) mutually interacting through their hydrophobic surfaces created by the protein chains when in α-helix conformation. The chains in each dimer are colored with gradient red/blue (Nt = solid color and Ct = faded color). Right: sphere representation of the hydrophobic (green) and hydrophilic (orange) surfaces of the apoA-I tetramer. (B) The crystal structure of the Δ43–apoA-I (Nt truncated) dimer (PDB id: 1AV1). Left: Proline residues that initiate 11 and 22 residue repeats are shown as yellow spheres. Right: the hydrophilic/hydrophobic surface of the dimer in sphere representation. The Ct domains of apoA-I chains have their hydrophobic surface facing outside the closed-loop dimer, while the helix-5 domains have their hydrophobic surface facing inside the dimer. (C) Left: Cartoon representation of the crystal structure of the Ct truncated form of lipid free apoA-I (1–185; PDB id: 3R2P). The Nt is shown to fold back and interact with the rest of the chain. Right: the hydrophilic/hydrophobic surface of the dimer is color coded with green/orange, respectively. The Nt domains of apoA-I chains have their hydrophobic surface facing toward the central region of the dimer, while the helix-5 domains have their hydrophobic surfaces facing each other.