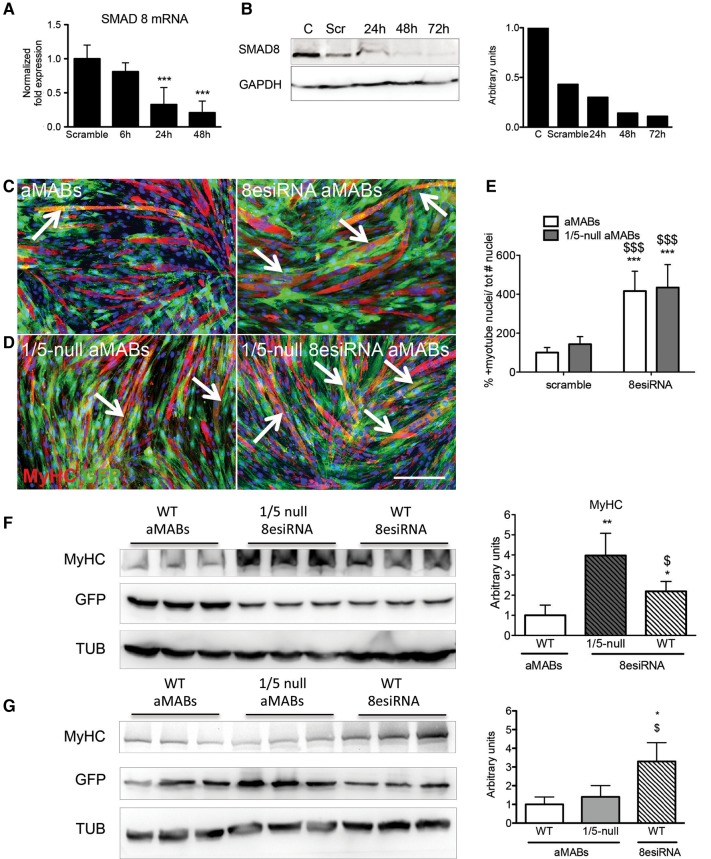
Figure 5.
Smad8 silencing by esiRNA in aMABs. (A) RT-qPCR analysis of Smad8 transcript at 6, 24, and 48 h after Smad8 silencing in Smad1/5-null aMABs. Data are expressed as fold expression compared with scramble treated cells and normalized to specific housekeeping genes (Gapdh, Hprt, and Tbp). Values are reported as mean ± SEM. (B) WB (left panel) and quantification (right panel) of SMAD8 protein levels normalized by GAPDH in Smad1/5-null aMABs silenced for Smad8 for 24, 48, and 72 h. (C) IF analysis for MyHC (red) and GFP (green) in WT aMAB co-cultures (left panel) and WT aMAB silenced for Smad8 co-cultures (indicated as 8esiRNA aMABs, right panel). (D) IF analysis for MyHC (red) and GFP (green) in Smad1/5- null aMAB co-cultures (indicated as 1/5-null aMABs, left panel) and Smad1/5-null aMABs silenced for Smad8 co-cultures (1/5-null 8esiRNA aMABs, right panel). MyHC and GFP double-positive myotubes are highlighted with arrows. (E) Fusion index of chimeric myotubes generated from Smad1/5-null aMABs shown in C and D is reported as percentage with respect to scramble treated aMABs. (F) WB (left panel) and quantification (right panel) of MyHC and GFP levels normalized by TUB in WT aMABs, Smad1/5-null aMABs + Smad8esiRNA (indicated as 1/5-null 8esiRNA), and WT aMABs + Smad8esiRNA (indicated as WT 8esiRNA). (G) WB (left panel) and quantification (right panel) of MyHC and GFP levels normalized by TUB in WT aMABs, Smad1/5-null aMABs (indicated as 1/5-null aMABs), and Smad8esiRNA-treated WT aMABs (indicated as WT 8esiRNA). Data are representative of three independent experiments and values are expressed as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 vs. C; $P < 0.05, $$P < 0.01, $$$P < 0.001 vs. respective controls. Scale bar, 500 μm.