FIGURE 1:
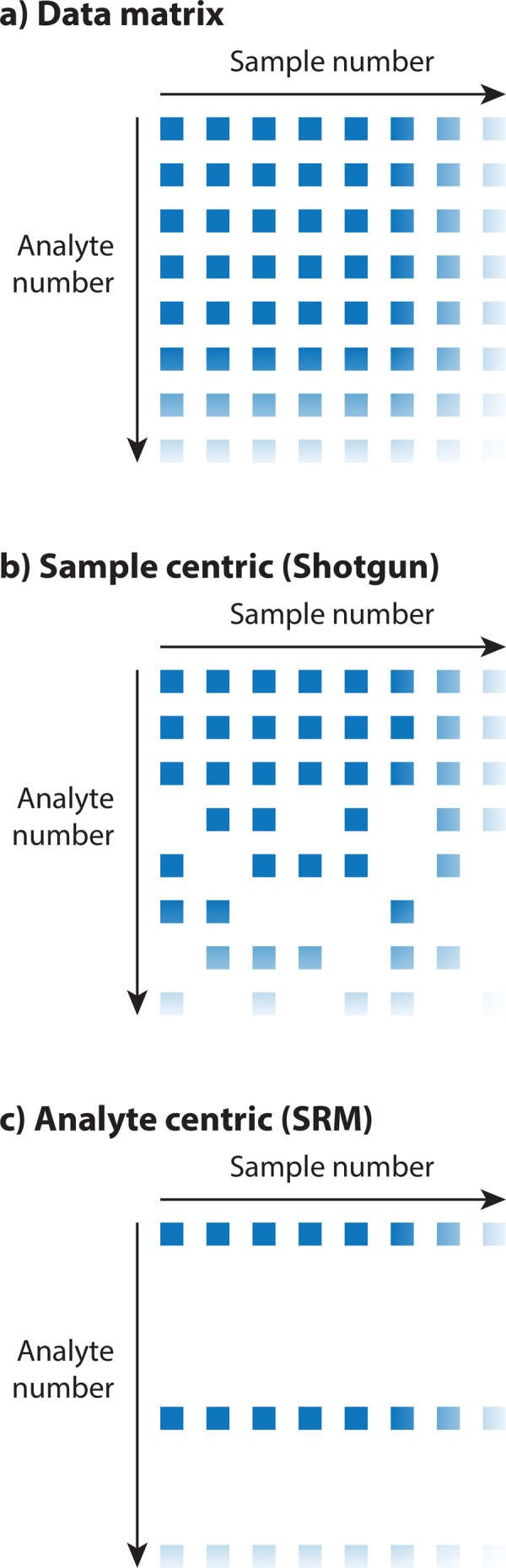
The proteotype data matrix as often found in proteomics experiments. (a) The data matrix contains quantitative values for different analytes (peptides or proteins) measured across multiple samples. One major goal in proteomics is to achieve high throughput (high number of quantified analytes) consistently quantified across many samples (experimental conditions, perturbations, or patient samples). (b) Sample-centric workflows (such as discovery proteomics or shotgun proteomics) place heavy emphasis on a high number of identifications in a single sample, which is achieved by data-dependent acquisition. However, the resulting data matrices often contain missing values due to undersampling issues, and in large studies, not all analytes can be quantified in every single sample. (c) In analyte-centric workflows (such as SRM and other low-throughput targeted proteomics techniques), the major focus is on achieving highly consistent quantification across many samples. The resulting data matrices are often devoid of missing values but only cover a few, carefully selected analytes.
