Studying signaling dynamics in single cells in vivo is critical to understanding how cells act and interact in 3D environments. Experimental and computational tools to quantify a circadian reporter in single cells in intact tissues for >1 wk are used to analyze the period, amplitude, and synchrony of circadian rhythms in vivo.
Abstract
Tracking molecular dynamics in single cells in vivo is instrumental to understanding how cells act and interact in tissues. Current tissue imaging approaches focus on short-term observation and typically nonendogenous or implanted samples. Here we develop an experimental and computational setup that allows for single-cell tracking of a transcriptional reporter over a period of >1 wk in the context of an intact tissue. We focus on the peripheral circadian clock as a model system and measure the circadian signaling of hundreds of cells from two tissues. The circadian clock is an autonomous oscillator whose behavior is well described in isolated cells, but in situ analysis of circadian signaling in single cells of peripheral tissues is as-yet uncharacterized. Our approach allowed us to investigate the oscillatory properties of individual clocks, determine how these properties are maintained among different cells, and assess how they compare to the population rhythm. These experiments, using a wide-field microscope, a previously generated reporter mouse, and custom software to track cells over days, suggest how many signaling pathways might be quantitatively characterized in explant models.
INTRODUCTION
Studies of the dynamics of proteins in single cells have revealed the behavior and heterogeneity of many key signaling pathways (Locke and Elowitz, 2009; Purvis and Lahav, 2013). The behavior of signaling molecules and the extent of variation among cells when they are organized into tissues and organs are rarely explored, mainly due to the complexity of studying protein dynamics in live tissues. Approaches to bridge this gap have used organoid systems and tissue slices—particularly neural slices—to study the effects of cell identity and environment on signal transduction (Gogolla et al., 2006; Williams et al., 2013). Although our understanding of tissue architecture and spatial regulation of signaling within tissues is considerable, it usually represents a static view with little information about fidelity or how signals might propagate among cells over time. The recently developed intravital imaging approach has improved our ability to analyze live cells within tissues, although it has been mainly used to study nonendogenous structures and is limited to an observation time of several hours (Pittet and Weissleder, 2011; Ellenbroek and van Rheenen, 2014). Extending this picture requires long-term observation at single-cell resolution of tissues expressing florescent reporters of a type that has thus far been largely limited to neural slices (Cheng et al., 2009).
There are multiple unique challenges to imaging single cells in live tissues. Direct in situ observation of individual cells in a tissue context requires imaging environments that offer ultrastable culture conditions while permitting constant observation. In addition, the complexity of analysis in the crowded and often highly autofluorescent environment requires sophisticated computational approaches to correct for aberrations and permit exposures that minimize the photon dose to the sample.
One system that has drawn particular attention in the efforts to move toward near–in vivo imaging is the circadian clock. Most organisms have circadian clocks—molecular oscillators that drive rhythmic processes in physiology and behavior (Gachon et al., 2004). In mammals, circadian clocks are cell autonomous and built on a transcription-translation negative feedback loop in which the transcription factor BMAL1-CLOCK drives the rhythmic expression of its own inhibitors, PER and CRY (Takahashi et al., 2008). Circadian clocks are found in the cells of most organs (Yamazaki et al., 2000), and the circadian clock in each organ drives a unique subset of rhythmic processes. Decades of work have empirically defined the molecular components of the circadian clock and how these components combine to construct robust oscillations (Gekakis et al., 1998; Lee et al., 2001; Preitner et al., 2002; Busino et al., 2007; Godinho et al., 2007; Siepka et al., 2007; Asher et al., 2008; Duong et al., 2011; Padmanabhan et al., 2012; Lande-Diner et al., 2013; Kim et al., 2014)
The activity of circadian clocks in peripheral organs has been studied extensively at the population level, mainly by measuring bioluminescent reporters of circadian transcription in cultured organ explants (Yamazaki et al., 2000). At present, there is no quantitative information about the dynamic behavior of circadian clocks at the single-cell level in any intact peripheral tissue. Here we develop experimental and computational approaches to quantify and analyze the dynamics of circadian clocks in live mouse tissue explants at the single-cell level and to characterize the diversity of the clock dynamics among cells and between pulses within a cell. Long-term imaging of individual circadian clocks in the context of tissue explants, as we develop and demonstrate here, will facilitate the characterization of the individual properties of each clock, its organization, and its contribution to the overall population rhythm. More globally, our approach can be used to study similar complex dynamical systems in which the overall output is derived from the integration of many individual signals.
RESULTS AND DISCUSSION
Circadian rhythms of Per1-YFP can be measured and quantified in tissue explants
We developed a system that allows long-term imaging of the circadian clock in individual cells in the context of their tissue of origin. We explanted tissues from a previously described transgenic mouse ubiquitously expressing Per1-YFP (YFP, yellow fluorescent protein), a validated fluorescent reporter of circadian clock activity (Cheng et al., 2009). For the purpose of this study, we chose to focus on the activity of clocks in osteocytes from the calvarial bone and tenocytes from the tail tendon. In these tissues, the relatively large distance between nuclei eliminates the need to deconvolve neighboring cells, and the nature of the extracellular matrix restricts cellular motility (Figure 1, A and B). The long observation times and the limited axial resolution of our microscopy was well suited to the relatively immobile and coplanar cells in the bone and tendon, enabling long-term analysis that would be challenging in other tissues. Microscopy modalities that provide greater three-dimensional imaging depth such as light sheet microscopy might make analysis of other tissues more tractable.
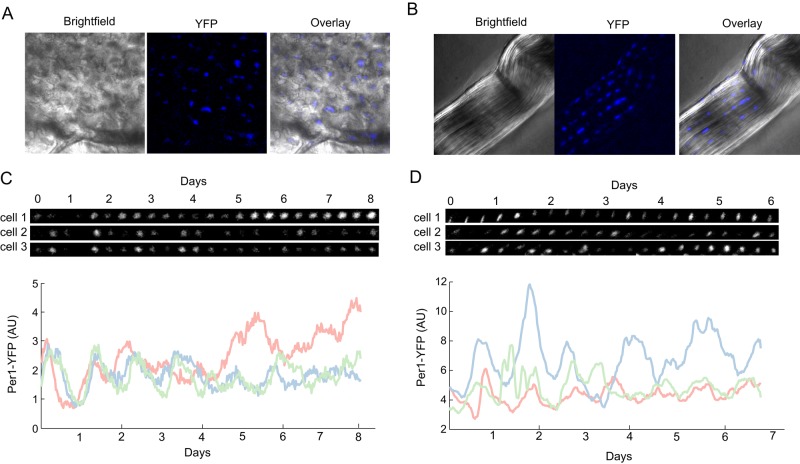
FIGURE 1:
A Per1-YFP reporter allows for single-cell quantification of circadian rhythms in a mouse organ explant system. (A, B) Bright-field and fluorescence images of bone from the calvarium (A) and tendon from tail (B). (C, D) Images and quantification of three bone (C) and three tendon (D) cells. Cells were imaged every 30 min. Frames every 8 h. Quantification of YFP intensity over time shows an oscillatory pattern with a periodicity of ∼24 h.
We cultured explanted tissues in a temperature-, CO2-, and humidity-controlled microscope, using a B27-supplemented transparent medium that maintained cell viability for >1 wk. This approach allows for single-cell imaging and long-term observations of tissue pieces in a near-natural tissue environment. Using this setting, we were able to image tissues for 6–8 d, sampling >40 fields of view every 30 min. Acquisition time per each field of view was <5 s, much faster than typical luciferase-based single-cell imaging, which requires at least several minutes (Welsh and Noguchi, 2012).
We acquired bright-field images of bone and tendon explants that capture the unique architecture of each tissue. Osteocytes were well separated and regularly distributed across the tissue surface (Figure 1A), and tenocytes (Figure 1B) were arranged in stripes. We measured the fluorescence intensity of Per1-YFP in individual cells over time and observed oscillations in both tissues (Supplemental Movies S1 and S2). Visual analysis of the traces suggested an oscillatory pattern with a period of ∼24 h (Figure 1, C and D), as expected from a circadian signal.
Individual cell oscillators have stable periods in the circadian range
We then collected data from five separate mice and developed software that identifies cells within each image, quantifies the YFP intensity, and strings images together to form tracks of single cells over days of observation. We recorded and quantified the levels of Per1-YFP in >600 cells from the calvarial bone and ∼150 from the tendon. Inspection of the extracted single- cell traces of the circadian dynamics in both tissues indicates that cells show pronounced oscillations in the expected circadian range (Figure 2, A and B). Circadian oscillations of similar characteristics were previously observed in single-cell traces from suprachiasmatic nuclei slices (Liu et al., 2007), as well as from dissociated fibroblasts (Welsh et al., 2004; Bieler et al., 2014).
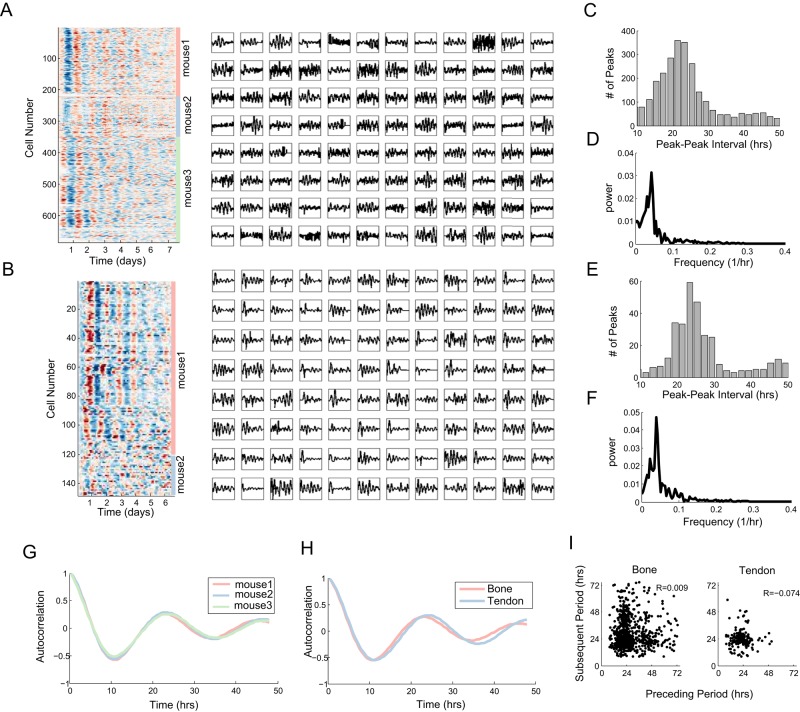
FIGURE 2:
Long-term time-lapse imaging and automated segmentation and analysis allow for large-scale acquisition of single-cell circadian data in tissues. (A, B) Heat map and representative line plots showing Per1-YFP oscillations in calvarial cells (A) and tendon (B) drawn from three (A) or two (B) mice. (C–F) Distributions of peak-to-peak intervals and frequencies for calvarial cells (C, D) and tendon cells (E, F). (G) Average autocorrelation of Per1-YFP signal in three mice shows near-identical period and decay rate (N = 343, 129, and 221). (H) Comparison of calvarial (N = 693) and tail tendon (N = 108) signals shows similar average autocorrelation. (I) Scatter plots showing that the length of each individual period is independent of the preceding period (p = 0.78, 0.31 by Student's t test).
Visual analysis of the single-cell traces suggests that cells exhibit synchronous circadian behavior at the beginning of the experiment, as indicated by the distinct stripes of signal in the heat map, but lose coherence as the experiment advances. This was true for both tissues tested. The initial synchrony we observed could represent the default state in the intact living tissue or a resetting effect of postmortem extraction and mounting of the tissue. The latter involves growth serum shocks and temperature fluctuations, both strong circadian-synchronizing agents (Balsalobre et al., 1998).
Analysis of the intervals between subsequent peaks revealed a strong circadian peak of approximately 24 h and substantial variability in the waveform of the traces, with a long tail and a small peak at roughly 48 h, suggesting either that some cells either skip a period or we fail to detect it (Figure 2, C and E). To quantify the periodicity in the YFP signal, we computed the Fourier transform from the pooled data of each organ. Reassuringly, using either the mode of the peak-to-peak intervals or the Fourier transform, we obtained a period of between 22 and 25 h for both tissues (Figure 2, D and F). These values are within the range of those typically reported for bioluminescence population circadian rhythms (Yamazaki et al., 2000; Yoo et al., 2004).
We then considered whether there is mouse-to-mouse variability in the oscillatory period or dephasing of the circadian signal. Comparing data collected on the calvarial bone for three different mice, we observe negligible differences in the autocorrelation curves, arguing that the measured period and dephasing are highly reproducible (Figure 2G). Similarly, comparison of the autocorrelation of the osteocytes and tenocytes from either the calvarial or tail tendon show a slightly lengthened period in the tendons but overall similar behaviors across these tissues (Figure 2H). The autocorrelation curves (Figure 2, G and H) suggested a period of 23 h, consistent with our other analyses (Figure 2, C–F).
To test for potential sources of systematic error in our measurements, we adapted a method developed to compare a measured circadian regulated activity—such as movement in animals—measured with some presumed error with the underlying clock fidelity by asking whether the sequential periods are negatively correlated in length (Pittendrigh and Daan, 1976a). A negative correlation suggests that the measurement has less fidelity than the underlying clock. Reassuringly, in neither the tenocyte nor the osteocyte data do we observe a significant negative correlation (Figure 2I; t test, p > 0.05).
Circadian signaling fidelity drops over the course of the experiment
Given that we acquired measurements across at least 7 d of oscillations for the osteocytes, we next asked whether the period and phase change over time. We compared the mean peak–peak interval for the first and last day of observation. We found that whereas the mean itself was not statistically different, the variability was greatly expanded by the eighth day of the experiment (Figure 3A). One possible explanation for this is that the fidelity of the oscillator begins to break down in some cells. Alternatively, the computational signal estimates may become more difficult as the tissue begins to decay. Consistent with a drop in fidelity and a general loss of synchrony, as implied by the autocorrelation function, the phase distribution of osteocytes expands monotonically as the experiment progresses (Figure 3B).
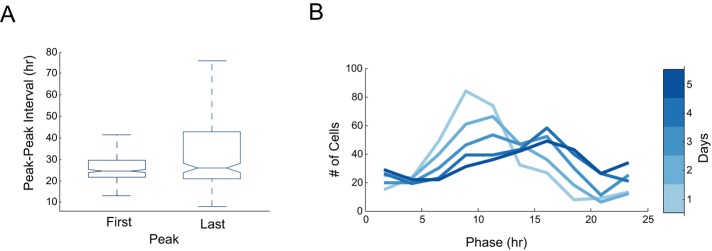
FIGURE 3:
Circadian signals in tissues show an initial synchrony that decays over time. (A) Period, measured by peak–peak interval, was calculated for the first and last circadian periods. Note that both measurements have comparable median values, but the last period has a wider variation. (B) The phase distribution of cells from the calvarial bone is plotted after 1, 2, 3, 4, and 5 d (for both A and B, N = 334 cells).
Variation in circadian amplitude is constrained within a cell and unrelated to period
The amplitude of circadian signaling has typically achieved less interest and study than the period. This is in part due to ease of measurement and also a lack of studies using single cells with uniform integrated reporter constructs. We quantified the amplitude of each oscillation at its peak for the osteocyte data and found a distribution of intensities ranging over roughly eightfold (Figure 4A). Further, unlike the period, we found that the variability of the amplitude does not increase with time, as both the mean amplitude and its variability were preserved from the first to the last oscillation (Figure 4A, inset). We then compared the overall variability in amplitude to the internal variation in a single cell. Of interest, we found that, on average, individual cells show lower variability in amplitude between their peaks than the variability observed between peaks in all cells (Figure 4B). This suggests that intrinsic factors that regulate the amplitude of circadian oscillations are preserved in cells throughout multiple days. The potential role of extrinsic factors, such as the local environment, remains to be explored.
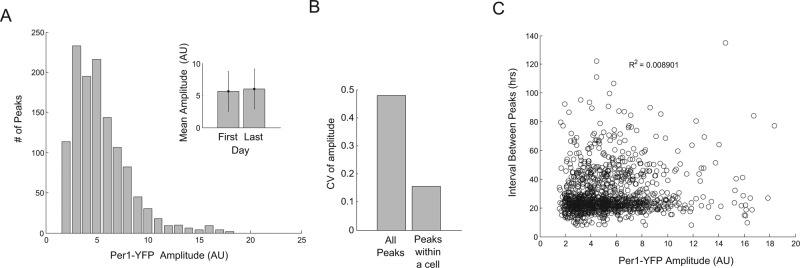
FIGURE 4:
The amplitude of Per1-YFP is constrained within a cell but varies across the population and is independent of the period. (A) Distribution of Per1-YFP amplitude across cells in the calvarial bone. Note that the average intensity of YFP across the population does not change over the duration of the experiment (inset). Error bars represent SD. (B) The coefficient of variation of Per1-YFP amplitude was computed across cells and within single cells across periods. The distributions are significantly different (Kolmogorov–Smirnov test, p = 10−54). (C) No correlation is observed between Per1-YFP amplitude and the period of the subsequent oscillation (for A–C, N = 254 cells).
Finally, we asked whether there was a relationship between period length and amplitude. We tested whether, for example, longer periods grant additional time for the Per1-YFP to accumulate, or, conversely, whether high circadian signaling might drive a faster period. Comparing the peak height to the subsequent interval between peaks, we find no substantial correlation (Figure 4C, Pearson r < 0.1), suggesting that in this system, period is buffered against variation in promoter activity of target genes. The conservation of period length among cells in a tissue is what one might expect if the system prioritizes the phase angles of individual cellular oscillators with respect to a common entrainment signal, much as precise period lengths of behavioral activity over time assure a consistent phase angle of activity with respect to the light–dark cycle (Pittendrigh and Daan, 1976b). In a tissue composed of weakly coupled cellular oscillators, this property would promote coherent and properly phased rhythmic function of the tissue as a whole.
MATERIALS AND METHODS
Tissue collection and sample preparation
Mice were housed in 12:12-h light/dark cycle and killed in accordance with a protocol approved by the Harvard Medical School Standing Committee on Animals. Organs were collected and immersed in ice-cold Hank's balanced salt solution supplemented with penicillin and streptomycin (Gibco, ThermoFisher Scientific, Cambridge, MA). Bone and tendon tissues were dissected using a Leica 2000 stereomicroscope to obtain ∼1 mm × 1 mm pieces. Explants were placed on MatTek 35-mm glass-bottomed dishes coated with 100 μl of 1.5% collagen (Cell Matrix, Hampshire, UK) and incubated for 5 min at 37°C. Fifty microliters of 1.5% collagen was added to cover the tissue, followed by a 10-min incubation at 37°C. Then 2 ml of transparent DMEM/F12 medium supplemented with penicillin, streptomycin, and B-27 (Life Technologies, Cambridge, MA) was added to each dish. The collagen coating immobilizes the tissue while allowing for nutrients and oxygen to penetrate. Calvarial-derived bone and tail tendons have a naturally thin cross section, and this feature enabled proper focusing of the objective on the cells. The depth of field of the microscope used was 3500 μm (see later description). When accounting for the thickness of the collagen layer, we found that to allow for proper visualization, samples needed to be in the range of 150 μm. We also found that tissues such as liver, in which cell density is high and nuclei are closely packed, were challenging to measure, as resolving individual nuclei proved to be difficult. Furthermore, tracking cells from tissues with high mitotic activity, such as lymph nodes, was difficult over the number of frames analyzed in one single experiment. In our experience, tissues in which nuclei are separated by an extracellular matrix constitute the best samples for this assay, as nuclei are well spaced and constrained in their movement. Additional tissues that meet these requirements and may be analyzed using similar approaches are adipose tissue, cartilage, and blood capillaries.
Time-lapse microscopy
Tissues were imaged using the VivaView system (Olympus, Waltham, MA). This system consists of a fully integrated and motorized inverted microscope with a cooled charge-coupled device camera that allows high-quality, long-term time-lapse imaging in a constant and optimized environment. The microscope has a rotating platform that can accommodate up to eight 35-mm dishes and allows for imaging of multiple positions in each one of the dishes. We used a magnification of 20× to capture the largest possible number of nuclei. Tissues were maintained in humid conditions (∼95% relative humidity), 37°C, and 5% CO2 for the entire imaging period. Differential interference contrast (DIC), Venus (Per1-YFP), and red fluorescent protein (RFP; autofluorescence) images were taken every 30 min for the duration of the experiment. Images were acquired using MetaMorph software (Molecular Devices, Silicon Valley, CA).
The microscope uses an X-Cite exacte fluorescence mercury lamp and a UPLSAPO20X objective (numerical aperture, 0.95; working distance, 0.18 mm). For the Venus filter set (enhanced YFP/Venus/citrine), excitation was 495 nm and emission was 540 nm. For the RFP filter set (Texas red/mCherry/Alexa Fluor 594), excitation was 560 nm and emission was 635 nm.
Image analysis
Images were processed and analyzed with custom Matlab (Mathworks, Natick, MA) code, which can be provided upon request. Briefly, each plane of a Z-stack was background subtracted, followed by maximum-intensity projection. Images were down-sampled twofold to improve signal. If substantial drift had occurred over the course of imaging, sequential time points were aligned using a local cross- correlation metric (Guizar-Sicairos et al., 2008). Images were then smoothed on the time axis using a Kalman filter. The first 5–10 h of data were then discarded as unreliable, as the tissue was often “settling” on the stage. Typically, 5–7 d of good quality data were obtained from a given experiment.
Each position was manually visualized in ImageJ (National Institutes of Health, Bethesda, MD) to determine whether it was of sufficient quality for further analysis. Components that went into this decision were number of visible cells, stability of the tissue over time (as accessed by DIC structure), and autofluorescence over time (as measured by RFP and YFP signal).
Nuclei were identified as objects with a given YFP intensity, shape, and size and tracked through subsequent frames, and intensity was computed as the mean of the brightest 10 pixels within an identified region. Identified cells were connected across frames using a nearest- neighbor algorithm resulting in single-cell traces; cell traces that did not persist for >80% of frames were discarded. Missing data (typically small numbers of frames in a trace in which a cell was missed by the automated analysis) was interpolated from a spline fit to the trace as a whole. Raw traces and the objects that gave rise to them were manually examined for correct tracking and cell-like properties (e.g., some degree of nonmonotonic change in intensity and visual verification that the cell track appeared continuous); typically, 30–50% of automated traces were discarded. Traces were then smoothed (averaging filter, width of 2.5 h) and baseline subtracted (48-h trend line was subtracted) for peak identification and period analysis; amplitude analysis used non–baseline-subtracted data.
Period was estimated by three methods: autocorrelation, peak-to-peak distance, and Fourier analysis. Briefly, autocorrelation was computed for each single-cell independently and averaged across all cells. Peaks were identified by smoothing the trace and identifying peaks using the inbuilt Matlab algorithm. Distance was then calculated for sequential pairs of peaks, and finally peaks from all cells were pooled to estimate the distribution of periods. The same peak positions were used to measure the peak amplitude. To estimate the Fourier transform, single-cell traces were Fourier transformed and the result averaged across cells; the peak of this was taken as the frequency.
Supplementary Material
Acknowledgments
We thank Adrian Granada for comments and suggestions throughout this project and all members of our laboratories for helpful discussion and suggestions. We thank Ming Liu for her technical help, the Neurobiology Department, the Neurobiology Imaging Facility, and Michelle Ocana for consultation and instrument availability that supported this work. This facility is supported in part by the Neural Imaging Center as part of National Institute of Neurological Disorders and Stroke P30 Core Center Grant NS072030. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation (L.L.-D, C.J.W.) and National Institute of Health Grant GM083303 (J.S.-O, G.L.).
Abbreviations used:
- DIC
differential interference contrast
- RFP
red fluorescent protein
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0403) on August 12, 2015.
REFERENCES
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bieler J, Cannaco R, Gustafson K, Gobet C, Gatfield D, Naef F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol. 2014;10:729. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SIH, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Cheng H-YM, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, Obrietan K. Segregation of expression of mPeriod gene homologs in neurons and glia: possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet. 2009;18:3110–3124. doi: 10.1093/hmg/ddp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek SI, van Rheenen J. Imaging hallmarks of cancer in living mice. Nat Rev Cancer. 2014;14:406–418. doi: 10.1038/nrc3742. [DOI] [PubMed] [Google Scholar]
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Galimberti I, DePaola V, Caroni P. Long-term live imaging of neuronal circuits in organotypic hippocampal slice cultures. Nat Protoc. 2006;1:1223–6. doi: 10.1038/nprot.2006.169. [DOI] [PubMed] [Google Scholar]
- Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kwak PB, Weitz CJ. Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol Cell. 2014;56:738–748. doi: 10.1016/j.molcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci USA. 2013;110:16021–16026. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Elowitz MB. Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol. 2009;7:383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–91. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. I. The stability and lability of spontaneous frequency. J Comp Physiol. 1976a;106:223–252. [Google Scholar]
- Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. IV. Entrainment: pacemaker as clock. J Comp Physiol. 1976b;106:291–331. [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–56. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Yoo S-H, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong H-K, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Noguchi T. Cellular bioluminescence imaging. Cold Spring Harb Protoc. 2012;2012:1028. doi: 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PR, Morgan JL, Kerschensteiner D, Wong RO. In vitro imaging of retinal whole mounts. Cold Spring Harb Protoc 2013, pdb.prot072645. 2013 doi: 10.1101/pdb.prot072645. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.