Fig. 2.
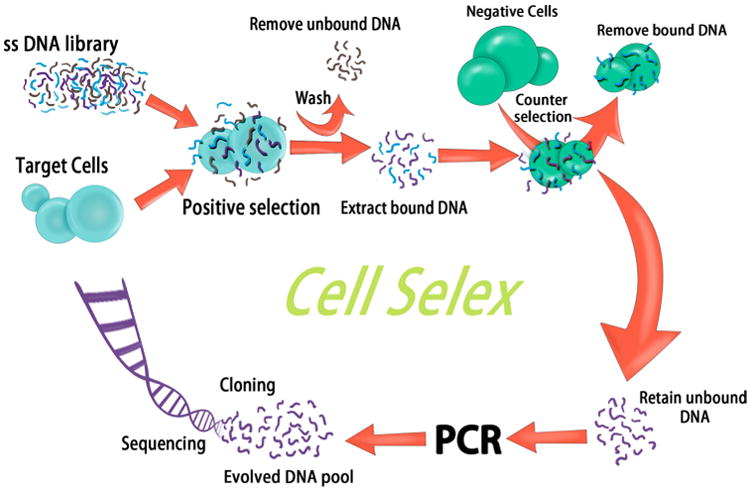
A schematic illustration showing the principle of cell-based systemic evolution of ligands by exponential enrichment (SELEX) for generation of aptamers based on their capability of recognizing complex molecular signatures of cancer cells rather than a single target on the cell membrane. Initially, a single-stranded nucleic acid (RNA/DNA) library is prepared (30–40 nucleotides in length flanked by primer sequences) followed by incubating the oligonucleotide library with a target cancer cell. The unbound DNA probes are washed out, and the bound ones are then collected and incubated with negative control cells for counter selection. All unbound probes are collected and amplified using polymerase chain reaction (PCR), and the evolved DNA pool is cloned and sequenced to determine the sequence of specific aptamers
