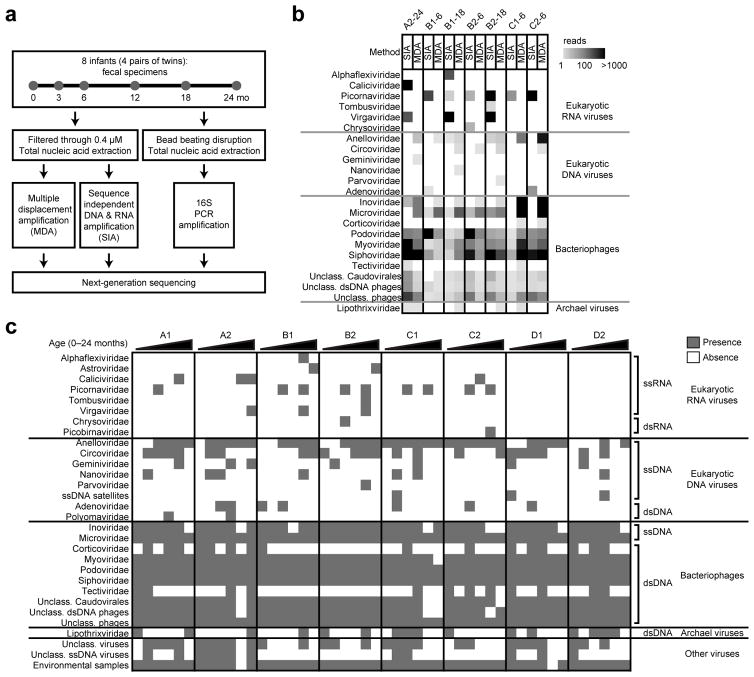
Figure 1.
Study design and metagenomic analysis of the infant gut virome. (a) Sequencing strategy to characterize the microbiome of 8 healthy infants (4 twin pairs). (b) Heatmap of reads assigned to virus families show that the profile is influenced by the sequencing method. Comparison of representative specimens is shown: fecal specimen from infant A2 at 24 months (A2-24), infant B1 at 6 months (B1-6) and 18 months (B1-18), infant B2 at 6 months (B2-6) and 18 months (B2-18), infant C1 at 6 months (C1-6) and infant C2 at 6 months (C2-6). SIA, sequence independent DNA and RNA amplification; MDA, multiple displacement amplification. (c) Presence-absence heatmap shows the viruses identified by subject (infants A1, A2, B1, B2, C1, C2, D1 and D2) and time point (0, 3, 6, 12, 18 and 24 months) during the first two years of life.