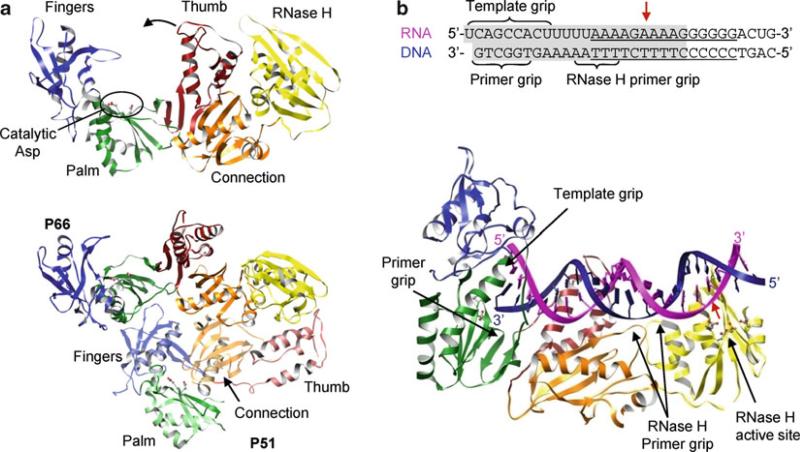
Fig. 12.11.
Structures of HIV reverse transcriptase. (a) p66 subunit (top) and p66–p51 heterodimer (bottom). The fingers (blue), palm (green), thumb (red), connection (orange), and RNase H (yellow) domains of p66 are in solid colors and domains of p51 are in transparent colors. Two aspartic residues in the YMDD motif (motif C) are shown as ball-and-stick models. The polymerase is in an “open” conformation due to binding of substrate, which is omitted in the figure for clarity. Note the connecting domain in p51 is inserted between the palm and the thumb domains, making p51 inactive as a polymerase. (b) Structure of p66–p51 heterodimer complexed with a PPT-containing RNA–DNA hybrid. Only the p66 subunit is shown for clarity. The sequence of the RNA–DNA hybrid used in the crystallization is shown with the PPT underlined and the portion visible in the crystal structure shaded in gray (top). Template RNA and primer DNA are shown in magenta and blue, respectively, and the RNase H cleavage site is indicated with a red arrow. The primer grip, template grip, and RNase H primer grip are indicated