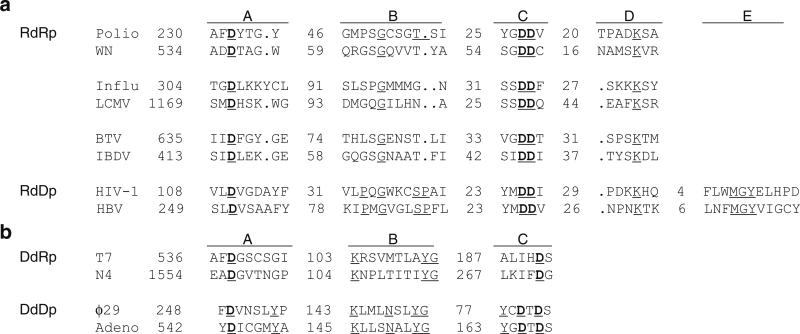
Fig. 12.3.
Polymerase sequence motifs in RNA-dependent and DNA-dependent polymerases. (a) Alignment of the RNA-dependent polymerase motifs A, B, C, D, and E. The alignment includes RdRps encoded by plus-sense ssRNA viruses (poliovirus and West Nile virus), minus-sense ssRNA viruses (influenza and Lassa viruses), dsRNA viruses (bluetongue and infectious bursal disease viruses), and DdRps (reverse transcriptases) encoded by a ssRNA virus (human immune deficiency virus type 1) and a dsDNA virus (hepatitis B virus). The numbers of residues separating the motifs are indicated. (b) Alignment of the DNA-dependent polymerase motifs A, B, and C. The alignment includes DdDps encoded by bacteriophages T7 and N4, and DdRps encoded by bacteriophages ϕ29 and adenovirus. DNA-dependent polymerase motifs A and C and RNA-dependent polymerase motifs A and C coordinate divalent metal ions for catalytic activity