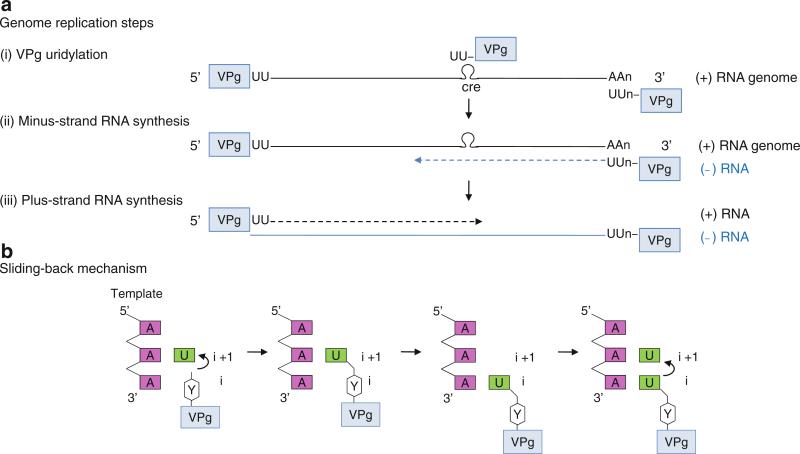
Fig. 12.5.
Poliovirus genome replication. (a) Genome replication steps. (i) VPg uridylation. Terminal protein VPg is covalently attached to the 5′-end of plus-sense viral genome. The hydroxyl group of VPg Tyr3 acts analogously to the free 3′-OH of the primer terminus to form an uridylated form of VPg, VPg-pU, which is then further elongated to VPg-pUpU. The VPg-pUpU can be synthesized from either the poly(A) tail or conserved cis-acting replication element (CRE) as a template. (ii) Minus-strand RNA synthesis. VPg-pUpU is used as a primer to base-pair with a plus-strand genome and elongated to form a VPg-linked minus-strand RNA (blue strand). (iii) Plus-strand RNA synthesis. The VPg-pUpU made using the CRE as a template is used as a primer for plus-strand RNA synthesis in the subsequent step. Thus, both plus- and minus-strand RNAs are VPg-linked at their 5′-ends in vivo. (b) Schematic of the sliding-back mechanism. The polymerase initiates VPg uridylation at the second A on the 3′-end template. The VPg-pU then slides back one base to base-pair with the 3′-terminal A, and the polymerase begins second uridylation reaction. Thus, the sliding-back mechanism requires two sequential adenosines in the template