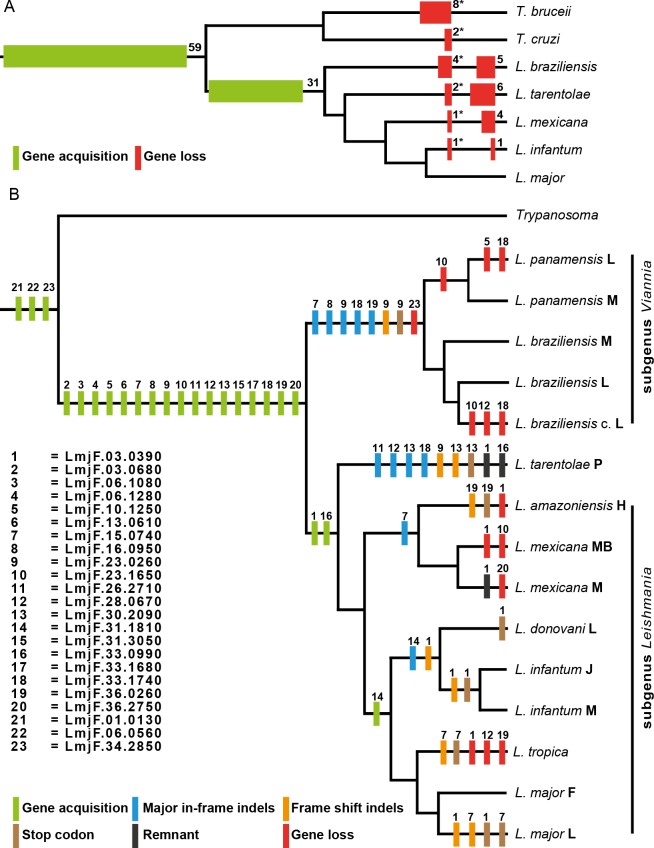
Fig 1. Consensus trees with orthologues to LGTs.
(A) Phylogenetic tree (congruent with the systematics proposed by Fraga and co-workers from 2010) showing the presence of LGT orthologs in five sequenced genomes of Leishmania and the sequenced genomes of Trypanosoma cruzi and Trypanosoma bruceii. The number of acquired genes (green boxes) and lost genes (red boxes) are presented numerically. (Red boxes*) refers to gene loss of orthologs to universal trypanosomatid LGTs. (B) Consensus tree summarizing the agreement between the phylogenetic trees for the twenty-three LGTs, investigated in detail in this study. Genus Leishmania is divided into the three subgenera Viannia, Sauroleishamania (here represented by only one species and one strain–L. tarentolae) and Leishmania. Subgenera Viannia and Leishmania are further divided into several complexes, and the topology of the consensus tree is completely congruent with the systematics proposed by Fraga and co-workers (2010). The fate of each LGT is illustrated with coloured boxes. “Gene acquisition” (green boxes) and “Gene loss” (red boxes) are based on presence of orthologs in published genomes or amplification of orthologs with PCR. Insertions/deletions (indels) (blue and orange boxes) and presence of stop codons (brown boxes) have been determined by manual inspections of alignments of obtained sequences and are relative to the gene sequence in L. major F. “In-frame Indels” (blue boxes) are insertions or deletions of more than 50 nucleotides not causing any frame-shift. “Frame-shift Indels” (orange boxes) are insertions or deletions that causes frame-shifts. Remnants (black boxes) of orthologs have been identified with synteny analyses (see Material and Methods).