Fig 3. LGTs with large variations in codon bias among their orthologs.
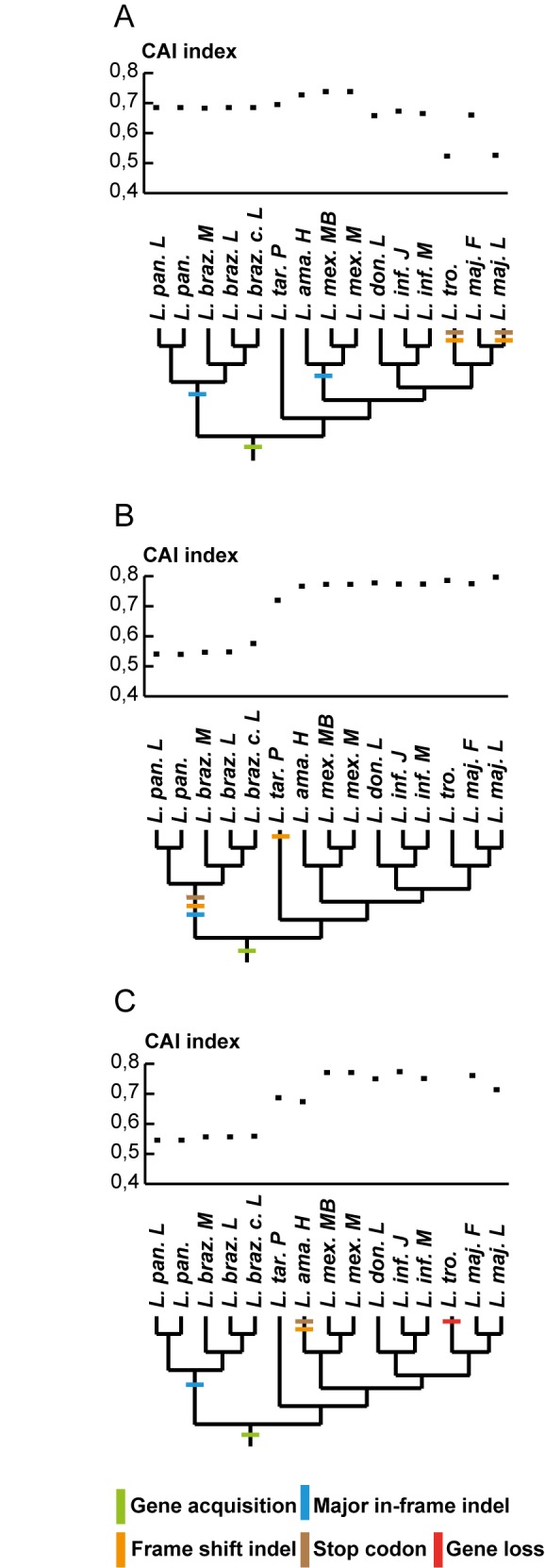
(A) CAI scores of orthologues to LmjF.15.0740, and the fate of the LGT in genus Leishmania. The average CAI score of LmjL.15.0740 and LtrX.15.0740 is significantly lower than the average CAI score of the other orthologs to LmjF.15.0740 (p<0,001). Frame shift indels and stop codons in LmjL.15.0740 and LtrX.15.0740, as well as major in-frame indels in subgenus Viannia and mexicana-complex are indicated in the phylogenetic tree. (B) CAI scores of orthologues to LmjF.23.0260, and the fate of the LGT in genus Leishmania. The average CAI scores of orthologs to LmjF.23.0260 in subgenus Viannia is significantly lower than the average CAI scores of the other orthologs to LmjF.23.0260 (p<0,001). Major in-frame indels, frame shift indels and stop codons in all orthologs of subgenus Viannia, as well as stop codon in L. tarentolae are indicated in the phylogenetic tree. (C) CAI scores of orthologues to LmjF.36.0260, and the fate of the LGT in genus Leishmania. The average CAI scores of orthologs to LmjF.36.0260 in subgenus Viannia is significantly lower than the average CAI scores of the other orthologs to LmjF.23.0260 (p<0,001). LamH.36.0260 displays the lowest CAI score among orthologs of subgenus Leishmania. Major in-frame indels contributing to divergence of orthologs of subgenus Viannia, frame shift indels and stop codons in LamH.36.0260 and gene loss in L. tropica are indicated in the phylogenetic tree. Significance in panel A-C was calculated with a pooled two-sample t-test.
