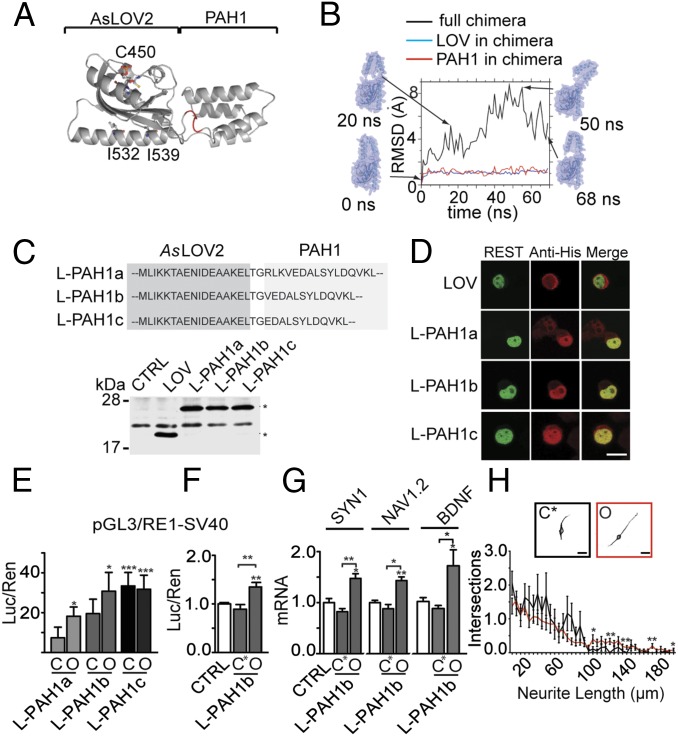
Fig. 2.
Building of the AsLOV2-PAH1 chimera. (A) Model of the AsLOV2-PAH1 chimera resulting from RosettaDock (35) and built using the LOV2 domain of A. sativa phototropin 1 (PDB ID code 2V1A) and the PAH1 domain of mSin3a (PDB ID code 2CZY). The backbone of AsLOV2 and PAH1 domains is represented as gray ribbons, whereas the LOV2 chromophore and the residues C450, I532, and I539 are represented as ball-and-stick. The linker amino acids are highlighted in red. (B) Conformational heterogeneity of the full chimera and its two domains along the MD simulation, calculated as RMSD from the starting structure. Large RMSD values for the full chimera correspond to highly distorted conformations generated because of the linker. Snapshots of conformations at different simulation times are also shown, with the molecular surface represented as transparent spheres. (C, Upper) Amino acid sequences of AsLOV2-PAH1 constructs, limited to the linker region. (C, Lower) N2a cells were transfected with a control plasmid (CTRL) or effector vectors encoding for AsLOV2 alone or AsLOV2-PAH1a, -b, or -c, as indicated. The expression of the constructs in total cellular lysates was analyzed by Western blot by using anti-histidine tag antibodies. Specific immunoreactive bands are indicated with asterisks. (D) Confocal images of N2a cells cotransfected with GFP-REST and either AsLOV2 alone or AsLOV2-PAH1a, -b, or -c, as indicated. N2a cells were processed for indirect immunofluorescence using anti-histidine tag antibodies (red) to detect AsLOV2 constructs or anti-EGFP (green) to detect GFP-REST. The overlay images (merge) reveal colocalization of REST and the AsLOV2-PAH1 constructs in the nuclear compartment. (Scale bar, 10 μm.) (E and F) HeLa cells were transfected with the pGL3-RE1/SV40 reporter vector in the absence (white bars) or presence (gray bars) of expression plasmids encoding for closed (C) or open (O) AsLOV2-PAH1a, AsLOV2-PAH1b, AsLOV2-PAH1c (E) or expression plasmids encoding for the closed double mutant (C450A-I532A) or open AsLOV2-PAH1b (F). Control samples were cotransfected with the empty vector corresponding to the effector plasmids. Luciferase activity was measured 48 h after transfection. Data were first normalized to the activity of the cotransfected TK-Renilla reporter vector and subsequently to the activity of the reporter gene alone, set to 1. (*P < 0.05; **P < 0.01 ***P < 0.001; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). Luc/Ren, luciferase/Renilla ratio. (G) Undifferentiated N2a cells were transfected with plasmids encoding for the closed double mutant (C450A-I532A) or open variants of the LOV2-PAH1b chimera, as indicated. The endogenous mRNA levels of SYN1, NAV1.2, and BDNF genes were quantified via qRT-PCR. GAPDH and HPRT1 were used as control housekeeping genes (*P < 0.05; **P < 0.01; one-way ANOVA followed by the Tukey’s multiple comparison test vs. control or the indicated group; n = 3 independent experiments). (H) Sholl analysis of N2a cells transfected with expression plasmids encoding for the closed (C) or open (O) mutants of AsLOV2-PAH1b and differentiated with Retinoic Acid (*P < 0.05; **P < 0.01; Student t test; n = 3 independent experiments). Representative cells are shown in the Insets. (Scale bar, 20 μm.) CTRL, control; LOV, AsLOV2; L-PAH1, AsLOV2-PAH1; L-PAH1b C*, AsLOV2-PAH1b (C450A-I532A).