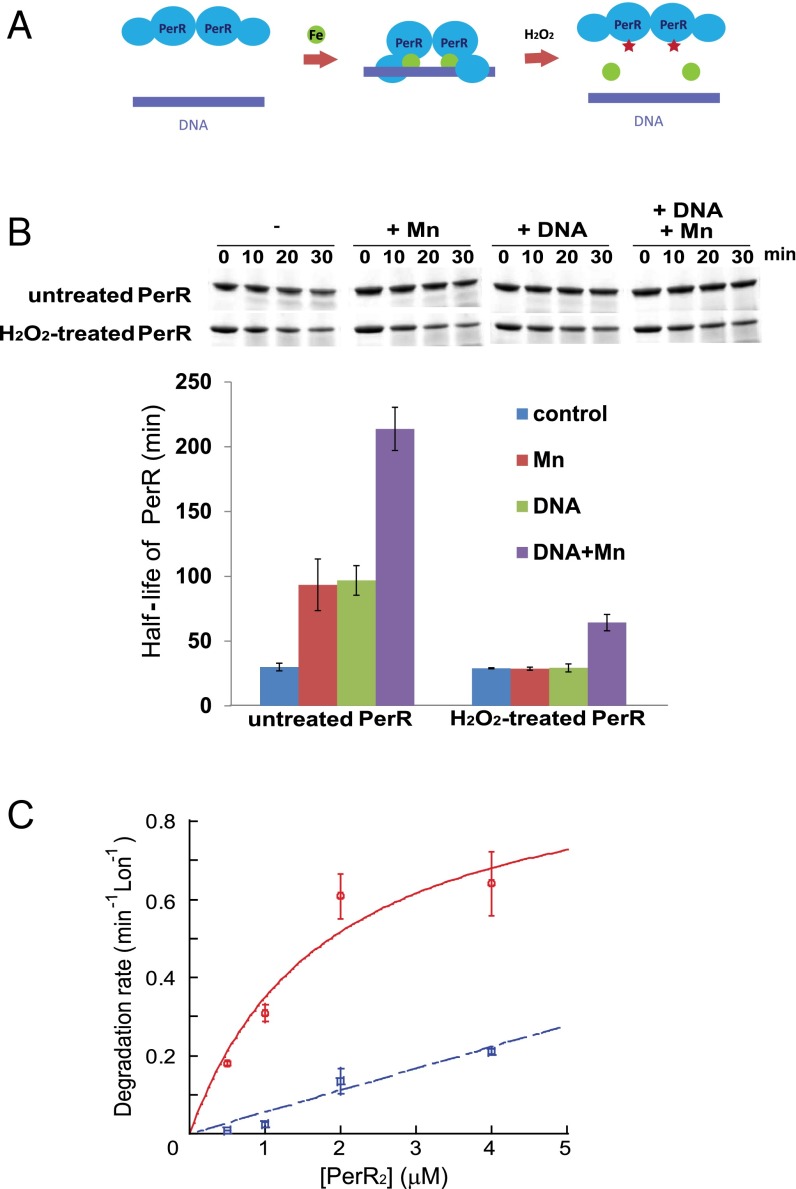
Fig. 3.
Binding of regulatory metal inhibits degradation of PerR by LonA. (A) Model for the PerR conformational changes induced/stabilized by regulatory metal-binding and oxidation by iron and H2O2. The green circles represent iron, and the red stars represent oxidized histidine residues at the sensor site. (B) In vitro degradation rates of untreated and H2O2-treated PerR by LonA were measured in the presence or absence of 100 µM MnCl2 and a 25-bp fragment of the mrgA promoter DNA, which carries a PerR-binding sequence (4). Upper is the representative figure of degradation reactions, and Lower presents the averaged PerR half-life from three independent experiments with ±1 SEM. (C) Manganese decreases productive interactions between PerR and LonA. Initial degradation rates for PerR by LonA at different concentrations of PerR in the absence (red circle, solid line) and presence (blue square, dashed line) of 100 µM MnCl2. PerR concentration values on the x axis are for the PerR dimer. Degradation rates of the PerR proteins were measured in the presence of 0.3 µM Lon monomer and 4 mM ATP. Plotted data represent the average value from three independent experiments with ±1 SEM and give a concentration for the half-maximal degradation (appKM) of apo-PerR by LonA of ∼1.9 ± 1.1 µM. The degradation rates were all normalized by dividing the observed rate by the concentration of Lon in the reaction (thus the rates shown are a hybrid between an initial velocity and a kcat value), and the rates are in units of PerR degraded per min, per Lon monomer.