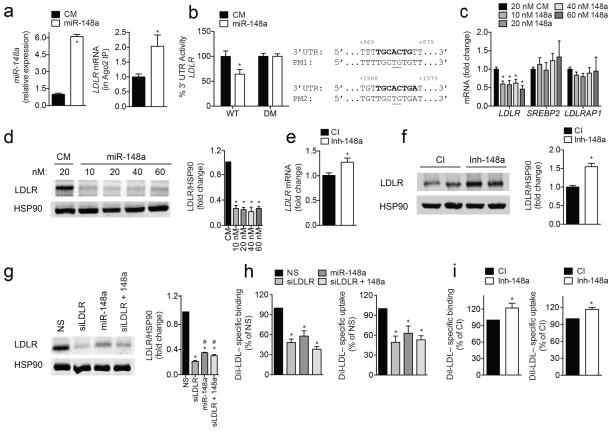
Figure 2. Post-transcriptional regulation of LDLR expression and activity by miR-148a in human hepatic cells.
a, qRT-PCR analysis of miR-148a and LDLR expression in Huh7 cells transfected with a control mimic (CM) or miR-148a mimic (miR-148a) after Ago2 immunoprecipitation. b, Luciferase reporter activity in COS7 cells transfected with CM or miR-148a and the human 3′ UTR of LDLR. Bold sequences, miR-148a binding sites; underlined nucleotides, point mutations (PM); WT, wild-type; DM, double mutation. c, d, qRT-PCR (c) and representative Western blot analysis (d) of LDLR in Huh7 cells transfected with CM or miR-148a. HSP90 was used as a loading control. e, f, qRT-PCR (e) and representative Western blot analysis (f) of LDLR in Huh7 cells transfected with a control inhibitor (CI) or inhibitor of miR-148a (Inh-148a). HSP90 was used as a loading control. g, h, Representative Western blot (g) and flow cytometry analysis of DiI-LDL binding and uptake (h) in Huh7 cells transfected with a non-silencing (NS) siRNA, miR-148a mimic, siRNA against LDLR (siLDLR) or both. *, P ≤ 0.05 compared to NS-transfected cells by one-way ANOVA. #, P ≤ 0.05 compared to siLDLR-transfected cells by one-way ANOVA with Bonferroni correction for multiple comparisons. i, Flow cytometry analysis of DiI-LDL binding and uptake in Huh7 cells transfected with CI or Inh-148a. In panels (a and c–i), data are the mean ± SEM of ≥ 3 experiments in duplicate. In panel (b), data are the mean ± SEM of ≥ 3 experiments in triplicate. *, P ≤ 0.05 compared to CM- or CI-transfected cells by unpaired t-test (a–f, i).