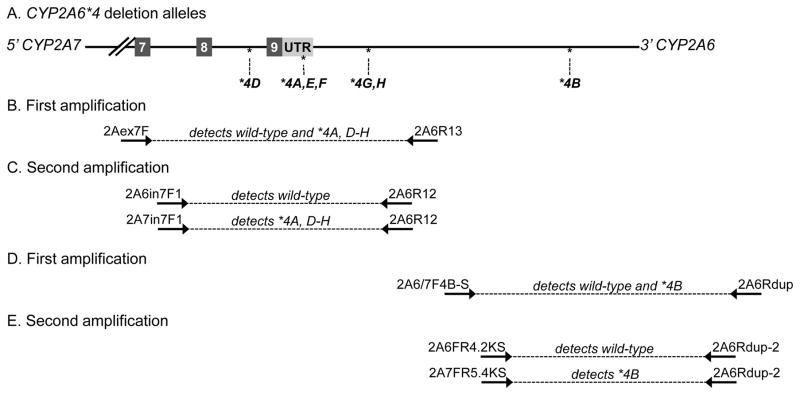
Figure 2.
CYP2A6*4 as an illustration of a CYP2A6-CYP2A7 hybrid allele. A. Crossover positions of the CYP2A6*4 alleles indicated relative to exons (numbered boxes). B. First amplification of PCR genotyping of CYP2A6*4H detects either wild-type CYP2A6 or all of the deletion alleles (except for *4B which is further downstream) using a 5′ primer that anneals to either CYP2A6 or CYP2A7, 2Aex7F, and a 3′ primer specific for CYP2A6, 2A6R13. C. Second amplification distinguishes between the wild-type allele and the deletion alleles using a 5′ primer that is specific to CYP2A6, 2A6in7F1, or to CYP2A7, 2A7in7F1, and a 3′ primer specific to CYP2A6, 2A6R12. D. First amplification of PCR genotyping of CYP2A6*4B detects either wild-type CYP2A6 or the *4B deletion allele using a 5′ primer that anneals to either CYP2A6 or CYP2A7, 2A6/7F4B-S, and a 3′ primer specific for CYP2A6, 2A6Rdup. E. Second amplification distinguishes between the wild-type allele and the *4B deletion alleles using a 5′ primer that is specific to CYP2A6, 2A6FR4.2KS, or to CYP2A7, 2A7FR5.4KS, and a 3′ primer specific to CYP2A6, 2A6Rdup-2. UTR: untranslated region