Fig. 2.
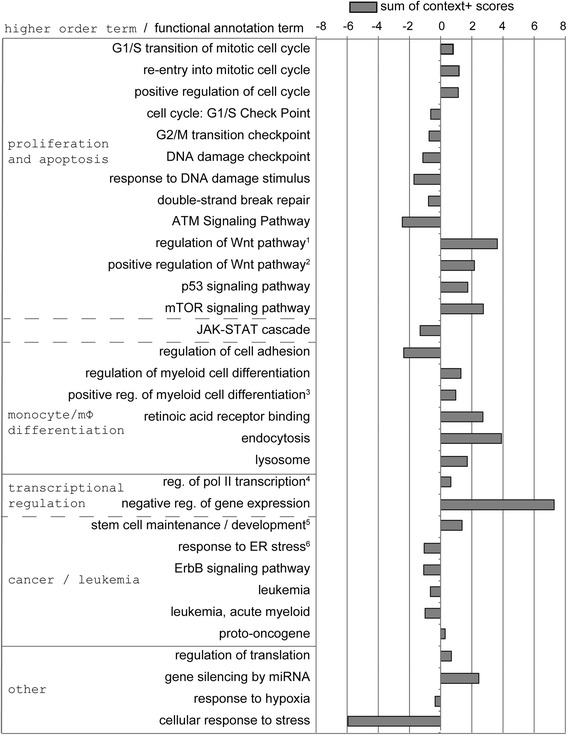
MiRNA-target gene prediction associated functional annotation terms identified via DAVID. Gene ontology analysis was performed for all THP1-expressed, targets predicted via TargetScan with a minimum context + score of ±0.4 of MLL-AF9 knockdown associated miRNAs. Annotation terms were manually assorted to five higher-order terms according to the major role of the process in the biological setting under investigation. Annotation terms assigned to two higher-order terms are placed between these two and separated by dotted division lines. Some annotations were abbreviated as indicated by superscript numbers: 1regulation of Wnt receptor signaling pathway, 2positive regulation of Wnt receptor signaling pathway, 3positive regulation of myeloid cell differentiation, 4regulation of transcription from RNA polymerase II promoter, 5stem cell maintenance / stem cell development (two synonymous terms), 6response to endoplasmic reticulum stress. Columns represent the sum of context + scores for all MTIs of this term, where MTIs of down-regulated miRNAs (in MLL-AF9 knockdown) retained negative context + score values, context + score values of MTIs of up-regulated miRNAs received positive values. Thus negative values of this column indicate strong predicted targeting by down-regulated miRNAs and positive values indicate strong predicted targeting by upregulated miRNAs, while targets which were similarly likely targeted by up- and down-regulated miRNAs do not reach the sum of context + score cutoff and are not included in the predicted target gene list
