Fig. 5.
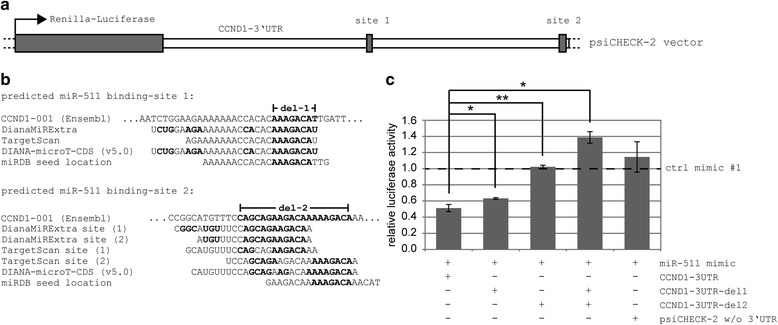
Effect of miR-511 on CCND1-3′UTR with and without deletion of miR-511 binding site seed regions. a Structural map of siCHECK2 vector: Wildtype full length CCND1-3′UTR consists of 3192 nucleotides and was cloned into the multiple cloning region located downstream of the Renilla luciferase translational stop codon of siCHECK-2 vector. Predicted miR-511 binding sites are indicated. b Sequence of miR-511 binding sites as predicted by the indicated algorithms. CCND1-3′UTR nucleotides complementary to miR-511 are indicated in bold. Positions 2738–2745 of CCND1-001 Ensemble transcript variant were deleted for binding site 1 seed region (del-1) and positions 4251–4270 for binding site 2 seed region (del-2). c Effect of miR-511 mimic on CCND1-3′UTR with and without seed region deletions in dual luciferase reporter assay. Columns represent mean over three independent experiments, bars indicate standard deviation; * p ≤ 0.05; ** p ≤ 0.005; w/o, without
