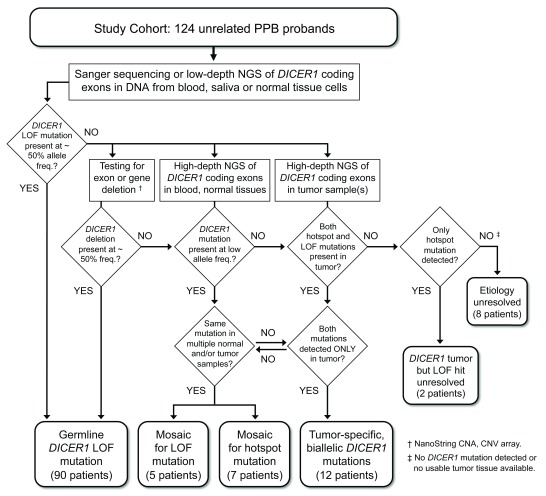
Figure 1. Study design – Detection and categorization of DICER1 mutations in PPB probands.
A cohort of 124 children diagnosed with pleuropulmonary blastoma (PPB) was screened for predisposing DICER1 mutations by targeted Sanger sequencing and/or low-depth, next-generation sequencing (NGS) of DNA amplified from peripheral blood cells, saliva (buccal cells) or non-neoplastic surgical specimens. Sequenced PCR amplicons covered the 26 coding exons of the DICER1 open reading frame and flanking splice signals. DICER1 coding sequence or splice site mutations detected at approximately heterozygous frequency in blood or normal tissue cells were categorized as germline mutations. For patients in whom screening revealed no germline mutation, blood and/or normal tissues were analyzed for the presence of intragenic deletions or larger genomic alterations using NanoString copy number assay and CNV array, and for coding or splice site mutations present at low allele frequencies using high-depth NGS on the Ion Torrent platform. Wherever possible, matched tumor specimens were also sequenced on the Ion Torrent platform. DICER1 mutations detected in tumor samples and at sub-heterozygous frequencies in blood or other normal tissue samples were categorized as mosaic mutations. RNase IIIb hotspot mutations detected in primary tumors of multiple organs were also categorized as mosaic mutations, even if they were not conclusively identified in blood or other normal tissues. Patients for whom both LOF and hotspot mutations were identified in a single tumor, but not found in blood or normal tissue samples, were categorized as having tumor-specific, biallelic DICER1 mutations. Cases of this last kind are considered sporadic PPB, not DICER1 syndrome.