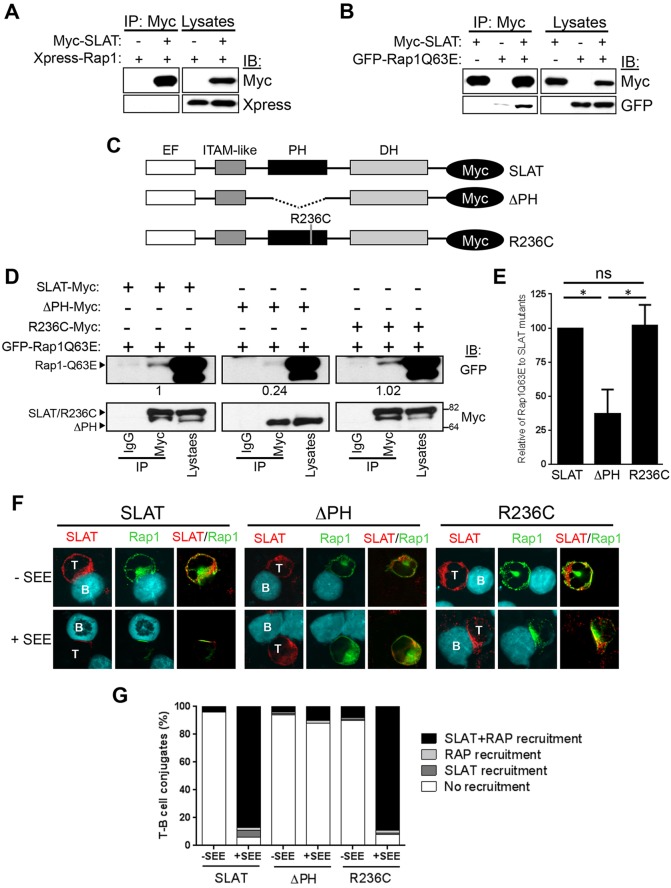
Fig. 3.
The PH domain of SLAT interacts with active Rap1 and is required to facilitate SLAT and Rap1 co-recruitment to the immunological synapse. (A,B) 293 T cells were co-transfected with Myc–SLAT and the indicated plasmids encoding Xpress–Rap1 (A) or GFP–Rap1Q63E (B). SLAT immunoprecipitation (IP) and cell lysates were analyzed by immunoblotting (IB). The results shown are representative of four independent experiments. (C) Schematic representation of Myc-tagged SLAT mutants. The EF hand, ITAM-like, PH and DH domains are shown. (D) 293 T cells were co-transfected with the indicated Myc-tagged SLAT plasmids together with GFP–Rap1Q63E. Cell lysates were immunoprecipitated with an anti-Myc antibody or normal IgG (as an immunoprecipitation control) and analyzed by immunoblotting with anti-Myc and anti-GFP antibodies. Cell lysates were also blotted with the same antibodies. Numbers under the Myc blots indicate the relative binding of Rap1Q63E to the SLAT mutants, as determined by densitometry. (E) Bar graphs representing the mean±s.d. relative binding of Rap1Q63E to SLAT mutants as determined by densitometry from three independent experiments as shown in D. (F) JA16 cells were co-transfected and stimulated as in Fig. 2C. Overlays of the red (SLAT) and green (Rap1) images are shown. B, B cell; T, T cell. The data are representative of three independent experiments. (G) Quantitative analysis of SLAT and Rap1 localization in the immunological synapse shown in F, representing approximately 100 T-cell–B-cell conjugates analyzed.