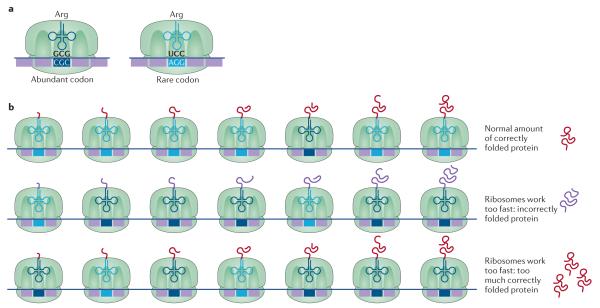
Figure 2. Biased codon usage.
a ∣ Biased codon usage occurs when synonymous codons are decoded by different tRNA isoacceptors. As some synonymous codons are used at higher frequencies than others, this leads to biased codon usage. b ∣ Although the optimized usage of synonymous codons increases the protein synthesis rate under most conditions, non-optimal codon usage can improve bacterial fitness under certain conditions. For example, codon optimization of the frq gene in Neurospora crassa results in a loss of fitness. frq controls the circadian clock function, and optimizing the codons of frq increases the expression of Frq but leads to defects in folding of this protein. These folding defects impair Frq function in the regulation of the circadian feedback loop (middle panel). Similarly, the kaiB and kaiC genes in the cold-adapted cyanobacterium Synechococcus elongatus are also enriched for non-optimal codons. The kaiB and kaiC genes are critical for regulation of circadian rhythms, and codon optimization increases the protein levels of KaiB and KaiC and prevents the circadian rhythm from switching off, resulting in loss of fitness by the cyanobacteria (bottom panel). Part b of the image adapted from REF. 144, Nature Publishing Group.