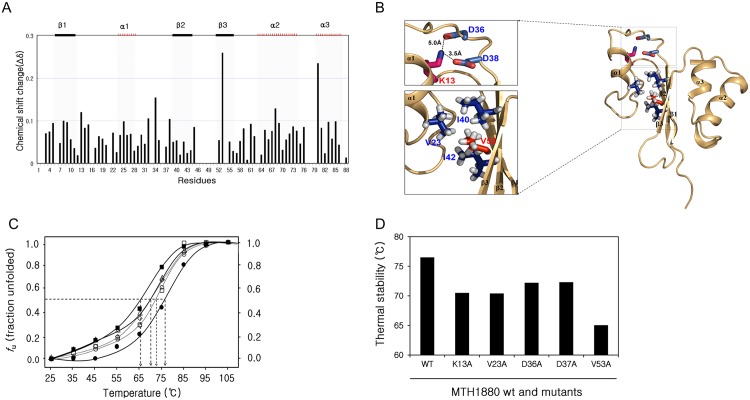
Fig 5. Residue-specific characteristics of MTH1880 unfolding.
(A) Chemical shift perturbation (CSP) analysis to detect residues susceptible to urea denaturation. The average chemical-shift changes were calculated using the following formula: ΔδAV = [(Δδ1H)2+(Δδ15N/5)2]1/2, where Δδ AV, Δδ 1H, and Δδ 15N are the average, proton, and 15N chemical-shift changes, respectively. (B) Structure of the MTH1880 represented by a ribbon diagram. K13-D36 and K13-D38 form salt bridges. Dashed lines indicate the salt bridges. K13-D36 and K13-D38 salt bridges contributed to the stability of the folding structure of MTH1880. Red and blue atoms mean oxygen and nitrogen, respectively. Hydrophobic core is formed by side-chain connectivity of hydrophobic residues. It is represented by sphere and stick, respectively. (C) Thermal-induced denaturation curves of wild type MTH1880 and mutants. The fraction of unfolding extracted from far-UV CD spectra at 222 nm with a constant heating rate of 10°C/h and 25μM protein concentrations. MTH1880 wt (filled circle), mutants in the salt bridge; K13A (triangle), D36A (square), D38A (circle) and mutants in the hydrophobic pocket; V23A (diamond), V53A(filled square). (D) Thermal stabilities were investigated for MTH1880 mutants. Tm values of mutant proteins are indicated as black bars.