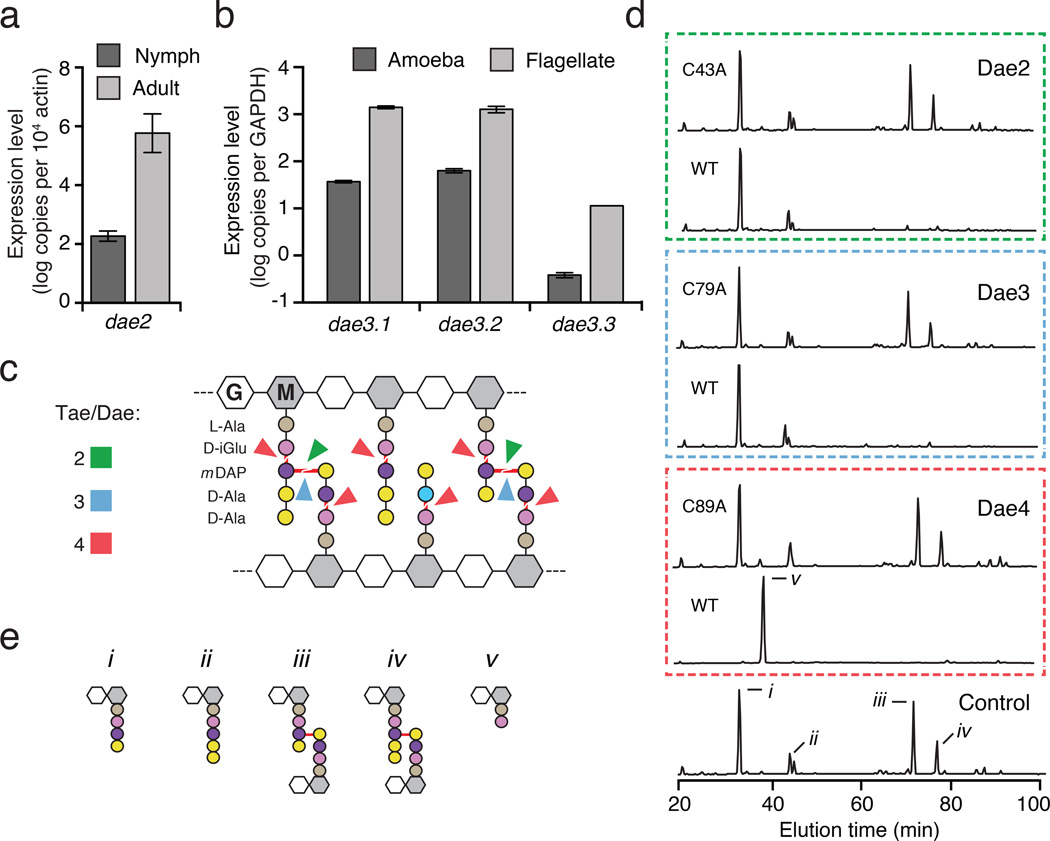
Figure 2. Eukaryotic dae genes encode differentially-expressed PG amidases with conserved specificity.
a, b, Expression profile of I. scapularis and N. gruberi dae genes at the indicated life stages as measured by qRT-PCR. Levels of each N. gruberi dae3 genes (Dae3.1–3.3) were determined. Error bars +/− s.d., n=3. c, Schematic representation of typical Gram-negative PG showing cleavage sites (red lines) for Tae and Dae families 2–4 (colors correspond to Fig. 1a). d, Partial HPLC chromatograms of E. coli PG sacculi products resulting from incubation with buffer (control), native and catalytically inactive (C43A, C79A, C89A) Dae enzymes and cellosyl. e, Major HPLC peaks assigned previously by mass spectrometry correspond to disaccharide-linked tetrapeptide (i), pentapeptide (ii), tetrapeptide– tetrapeptide (iii), pentapeptide–tetrapeptide (iv), and dipeptide (v)3.