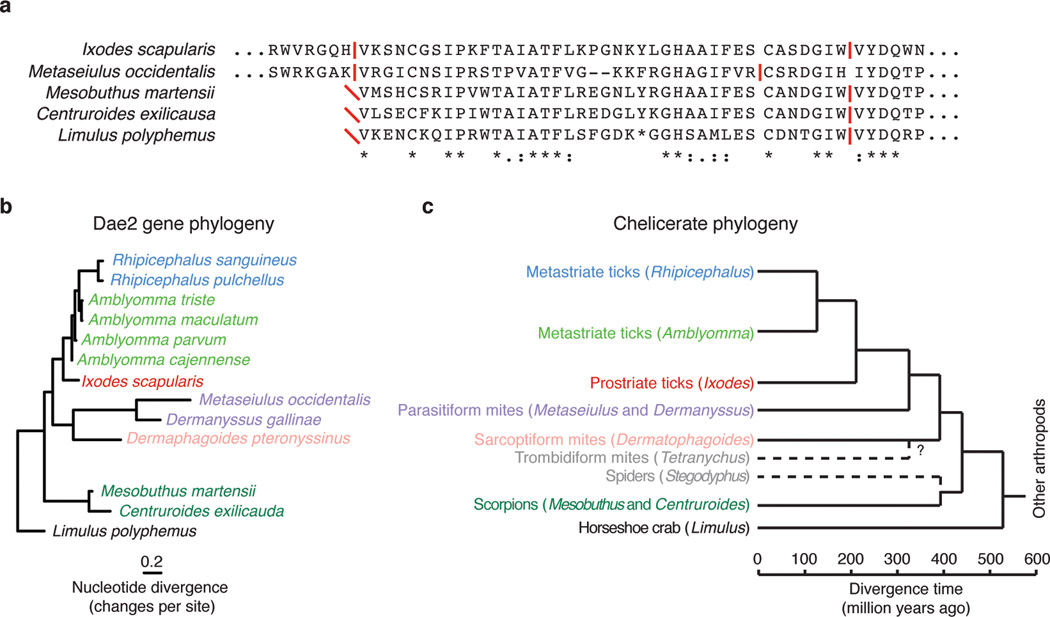
Extended Data Figure 5. Evidence for dae2 in other chelicerates.
a, Phylogenetic tree based on partial nucleotide sequences of dae2 from the indicated chelicerate species. Scale bar shows estimated divergence, in substitutions per nucleotide. b, Chelicerate phylogeny with approximate dates of divergence14. The unknown divergence time of sarcoptiform and trombidiform mites is indicated by a question mark. We find no evidence for dae2 in the complete genome of the trombidiform mite Tetranychus urticae nor in the partial (several species) or complete genome (Stegodyphus mimosarum) of any spider. Putative dae2 gene loss events in trombidiform mites and spiders are denoted (dashed lines). c, Alignment of Dae2 from ticks and mites (I. scapularis and M. occidentalis) with Dae2 sequences from partially assembled genomes of two scorpions (Mesobuthus martensii and Centruroides exilicauda) and the horsehoe crab (Limulus polyphemus). Splice junctions are denoted (horizontal red lines). All three alignable partial sequences start (red diagonal slashes) in the same position as the shared splice site in tick and mite dae2 genes, suggesting this is likely the beginning of the exons in all dae genes shown. A second intron position is shared between the tick, scorpion and horseshoe crab dae genes and is nearby the mite intron position.