Abstract
C/EBPα plays an important role in the modulation of cell proliferation, differentiation or apoptosis in various tissues. Most recently, reduced expression of C/EBPα and growth inhibitory effect was found in primary mammary carcinomas. However, the underlying mechanism is still not fully aware. Here, we firstly identified miR-134 as a target of C/EBPα in MCF7 breast cancer cell lines. C/EBPα overexpression promoted miR-134 expression, causing suppression of apoptosis- protective genes CREB and Bcl-2, and resulted in the proliferation inhibition of MCF7 cells. Moreover, anti-miR-134 rescued the proliferation inhibition of MCF7 cells and the suppression of anti-apoptotic genes CREB and Bcl-2 caused by C/EBPα overexpression. Collectively, C/EBPα inhibited cell growth in breast cancer cells via a novel pathway miR-134/CREB.
Keywords: C/EBPα, breast cancer cell, miR-134, proliferation, CREB
Introduction
Breast cancer is a common threat to women’s health. Originating in undifferentiated terminal structures of the mammary gland, breast cancer involves the clonal expansion of a transformed cell into an epithelial hyperplasia until the formation of adenocarcinomas [1]. Numerous transcription factors or other molecules were overexpressed or silenced during this process. Among the transcription factors, CCAAT/enhancer binding proteins (C/EBP) have been implicated in cellular proliferation or apoptosis in multiple tissues including mammary gland [2,3].
C/EBPα, the founding member of the C/EBP family, plays an important role in the modulation of cell proliferation, differentiation or apoptosis in various tissues, including adipose, liver, lung and blood [2,4-6]. Most recently, reduced expression of C/EBPα was found in primary mammary carcinomas. Overexpression of C/EBPα in breast cancer cell lines led to inhibition of proliferation [7]. However, the underlying mechanism is still not fully aware.
MicroRNAs are small non-coding RNAs that control the translation of target messenger RNAs, thus regulating various critical biological processes. MiR-134 was discovered as a brain-specific miRNA and it played an essential role in the differentiation of the embryonic stem cell to central nervous system by suppression of Nanog [8]. It was also reported that SIRT1 regulated memory and plasticity via miR-134-mediated post-transcriptional regulation of cAMP response binding protein (CREB), an anti-apoptotic gene in a number of tissues [9]. For a long time, miR-134 was thought to only act in nervous system [10,11]. However, much lately, a large-scale screening identified miR-134 as a candidate tumor metastasis suppressor, for its down-regulation in hepatocellular carcinoma (HCC) and significantly inhibitory effect on invasion and metastasis in HCC [12]. In addition, miR-134 was demonstrated to regulate epithelial-mesenchymal transition in lung adenocarcinoma cells [13]. These findings in non-nervous system suggested that miR-134 may play a role in more other tissues.
In this current study, we reported that miR-134 level was markedly distinct between normal and cancerous breast tissues. MiR-134 expression pattern was consistent with C/EBPα during the MCF7 breast cancer cell proliferation. Moreover, overexpression and silence of C/EBPα caused elevation and descent of miR-134 level respectively. Simultaneously, the anti-apoptotic gene CREB and B-cell lymphoma-2 (Bcl-2) were down- and up-regulated response to the alternation of miR-134 level. Collectively, we found a novel pathway that C/EBPα induced breast cancer cell apoptosis via miR-134/CREB.
Materials and methods
Samples
RNA was excreted from the mammary cells of 48 healthy and excised primary breast tumors of 48 women treated at Affiliated Xijing Hospital, Fourth Military Medical University, Xi’an, China, from 2012 to 2014. The samples were examined histologically for the presence of tumor cells.
Cell culture
MCF-7 breast cancer cells were purchased from the American Type Culture Collec-tion (Manassas, VA) and were cultured in DMEM supplemented with 10% FBS (Gibco, Invitrogen, Carlsbad, CA) and 1% penicillin, streptomycin, and neomycin antibiotic mixture. Cells were grown in a humidified incubator with an atmosphere of 95% air-5% CO2 at 37°C.
Transfection of oligonucleotides and plasmids
The pcDNA-C/EBPα expression plasmid was constructed and tested to be effective by Dingguo Changsheng Bio-tech-nology Co. LTD. (Beijing, China). Oligo antagonist and mimic for miR-134 were designed and synthesized by Ribobio Biotechnology Co. LTD. (Guangzhou, China).
For transfection, MCF7 cells were seeded into 6-well plates. When growing to 80% of confluence, their medium was changed to MEM without FBS and antibiotic mixture. After starvation for 4 hours, the medium was changed for DMEM supplemented with 10% FBS. Immediately, 6 μg of pcDNA-C/EBPα, or 100 nm miR-134 mimic, or 100 nm Oligo antagonist for miR-134 were co-transfected or transfected alone into the cells with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction. After another 6-8 h, the medium was changed for fresh DMEM supplemented with 10% FBS.
RNA extraction and RT-qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The resultant RNA was applied in the reverse transcription reaction to obtain the first chain cDNA. Real-time qPCR reactions were carried out in a final volume of 25 μl, using SYBR Premix Ex Taq (TaKaRa), 0.4 mM of each primer, and 200 ng of cDNA template. Each individual sample was run in triplicate wells. The reactions were initially denatured at 95°C for 3 min followed by 35 cycles of 95°C for 15 s, 60°C for 1 min. Quantification of amplicons was done using ABI 7300 software (Applied Biosystem). The primers applied in the reactions are as follows: C/EBPα (F: 5’-TGG ACA AGA ACA GCA ACG AG-3’, R: 5’-TTG TCA CTG GTC AGC TCC AG-3’), CREB (5’-TCA GCC GGG TAC TAC CAT TC-3’, 5’-TTC AGC AGG CTG TGT AGG AA-3’), Bcl-2 (5’-ATT GTG GCC TTC TTT GAG TTC G-3’, 5’-CAT CCC AGC CTC CGT TAT CC-3’), 18S (F: 5’-AAA CGG CTA CCA CAT CCA AG-3’, R: 5’-CCT CCA ATG GAT CCT CGT TA-3’). MiR-134 Stem-loop primer and quantitative primers were de-signed and produced by Ribo-bio Biotech.
Western blotting
Cells were washed with PBS, and lysed in NP40 lysis buffer (50 mMTris-HCl, 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, 10% glycerol) with protease inhibitors cocktail (Sigma). Proteins were separated in SDS-PAGE, transferred, and immunoblotted with various antibodies. The antibodies used were anti-C/EBPα (dilution 1:500; Abcam), anti-CREB (dilution 1:500; Cell Signaling), anti-Bcl-2 (dilution 1:500; Cell Signaling), and anti-GAPDH (dilution 1:5000; Sigma).
Web servers
The regulation relationship was predicted by the online server ChIPBase (http://deepbase.sysu.edu.cn/chipbase/), which is an integrated resource and platform for transcriptional regulation of long non-coding RNAs (lncRNAs), microRNAs, other ncRNAs and protein-coding genes based on ChIP-Seq data.
Statistical analysis
All data were expressed as mean ± S.E.M. of three or more biological replicates. Two-tailed Student’s t-test was employed to determine P-values. The significance was set at P < 0.05, and extremely significance at P < 0.01.
Results
C/EBPα and miR-134 were both down-regulated in cancerous breast tissues
Relative levels of C/EBPα and miR-134 in 48 normal breast tissues and 48 primary breast cancer samples were compared. The real-time qPCR analysis showed that the level of C/EBPα mRNA in primary breast cancer samples was just about 1/3 of that of normal breast samples (Figure 1A). Interestingly, we detected a similar expression pattern of miR-134. The level of miR-134 in the cancerous samples was about 1/2 of that of the normal ones (Figure 1B). C/EBPα and miR-134 were both down-regulated in cancerous breast tissues, hinting that there may be some association between C/EBPα and miR-134.
Figure 1.
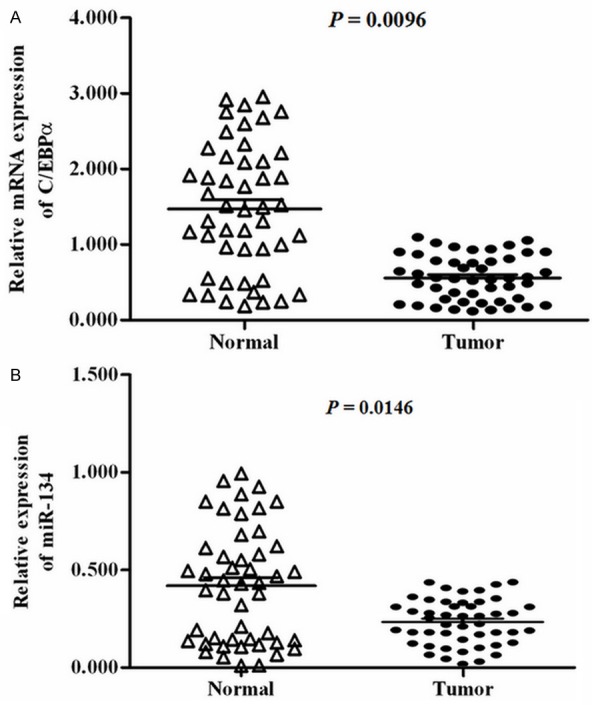
Expression of C/EBPα and miR-134 in normal and cancerous breast tissue. Relative expression levels of C/EBPα (A) and miR-134 (B) are shown in 48 normal breast tissues, 48 primary breast cancer samples. The relative level of C/EBPα was normalized so that the mean ratio of the 48 cancerous breast samples equals a value of 0.5. The relative level of miR-134 was normalized to the mean value of 18S RNA in the 48 normal breast tissues.
Mir-134 was promoted by in MCF-7 breast cancer cells
Then we tried to clarify the regulatory relationship between C/EBPα and miR-134. The predict output of the online server ChIPBase, an integrated resource and platform for transcriptional regulation of non-coding RNAs and protein-coding genes based on ChIP-Seq, showed that miR-134 was likely to be a target of C/EBPα (Figure 2A).
Figure 2.
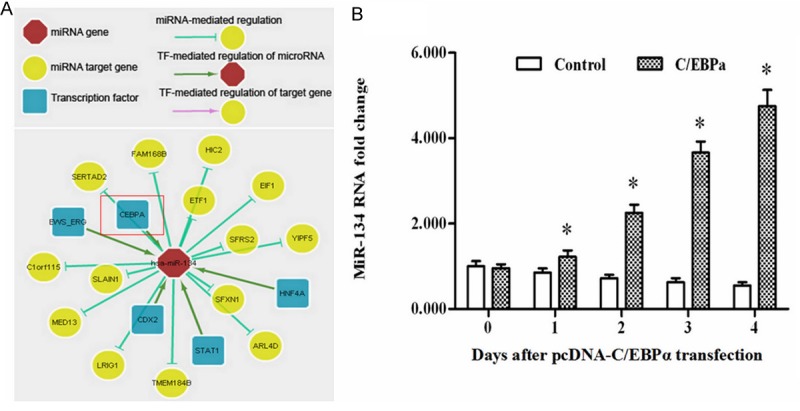
C/EBPα promoted expression of miR-134 in MCF-7 breast cancer cells. A. The regulation relationship MiR-134 is predicted to be promoted by C/EBPα using the online server ChIPBase. B. Overexpression of C/EBPα elevated the level of miR-134 in MCF-7 cells. The pcDNA-C/EBPα was transfected into MCF-7 cells. At time points of d 0, d 1, d 2, d 3, d 4, the level of miR-134 in the cells was determined by real-time qPCR. *P < 0.05.
The pcDNA-C/EBPα was transfected into MCF-7 cells. At time points of d 0, d 1, d 2, d 3, d 4, the level of miR-134 in the cells was determined by real-time qPCR. The data indicated overexpression of C/EBPα promoted miR-134 expression in MCF-7 cells (Figure 2B). CREB, target gene of miR-134, also an important anti-apoptotic gene in many processes, was decreased response to the increase of miR-134 (Figure 3A). Another apoptosis-inhibitory factor Bcl-2 was also minimized after transfection (Figure 3A). Simul-taneously, cell count analysis showed the cell proliferation was dramatically inhibited by C/EBPα overexpression (Figure 3B).
Figure 3.
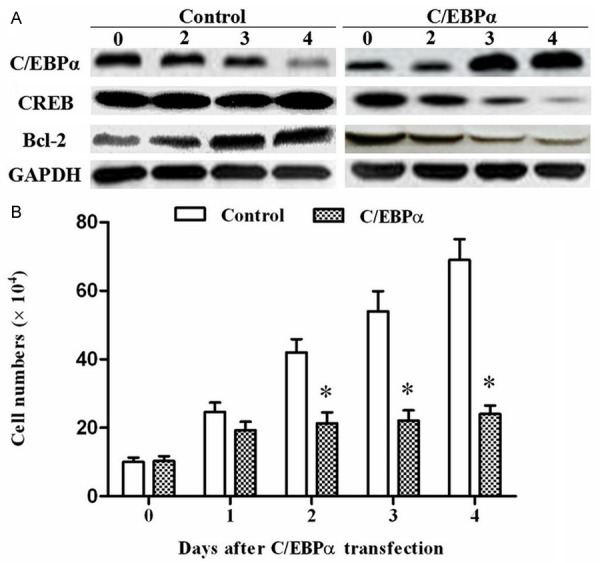
Overexpression of C/EBPα decreased cell proliferation of MCF-7 breast cancer cells. A. Overexpression of C/EBPα decreased expression of CREB and Bcl-2. At time points of day 0, day 2, day 3, day 4, expression of the anti-apoptotic genes CREB and Bcl-2 in the MCF-7 cells were determined by Western blotting. B. MCF-7 cell proliferation was decreased by C/EBPα overexpression. After transfection, the cell numbers were counted by Cellometer® Cell Counters and Cell Analysis Systems (Nexcelom Bioscience LLC, Lawrence, MA). *P < 0.05.
Anti-miR-134 rescued the proliferation inhibition by C/EBPα overexpression
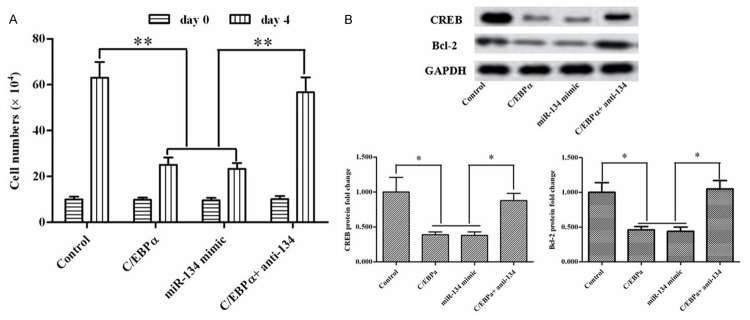
To testify the inhibition was associated with the elevation of miR-134. The pcDNA-C/EBPα, or miR-134 mimic, or miR-134 antagonist (anti-miR-134) were transfected alone or co-transfected into MCF7 breast cancer cell lines. At day 4 of transfection, the cell numbers were counted by the Cell Counters and Cell Analysis Systems. The result pointed that mimic miR-134 has the same effect with C/EBPα overexpression on inhibition of MCF7 cells and anti-miR-134 rescued the inhibition (Figure 4A). In consistent with the result of the cell count analysis, the suppression of anti-apoptotic genes CREB and Bcl-2 was also alleviated (Figure 4B). Therefore, C/EBPα inhibited cell proliferation via regulating miR-134/CREB.
Figure 4.
MiR-134 inhibitor rescued the proliferation inhibitory effect by C/EBPα overexpression. The pcDNA-C/EBPα, or miR-134 mimic, or miR-134 inhibitor were transfected alone or co-transfected into MCF7 breast cancer cell lines for 4 day. A. The cell numbers were counted by Cellometer® Cell Counters and Cell Analysis Systems. B. Expression of CREB and Bcl-2 proteins were detected by Western blotting. *P < 0.05.
Discussions
Although several reports have indicated that C/EBPα had an proliferation inhibitory effect in breast cancer [7,14]. However, not much is known about the underlying mechanism and network mediating the anti-proliferation impacts of C/EBPα. The present study showed that C/EBPα and miR-134 were consistently down-regulated in breast cancer and had an apoptosis-inducing effect in MCF7 breast cancer cell lines. This apoptosis-inducing effect was mediated by miR-134 and its target anti-apoptotic gene CREB.
C/EBPα was most well-known as an import regulator in adipocyte differentiation [5]. MiR-134 was thought to be nervous-specific for a long time. Though most recently a few evidence showed that it also functioned in HCC (Zha et al, 2014) and lung adenocarcinoma cells (Kitamura et al, 2014), there still is no report that it has an effect on development of breast cancer. In this current study, we showed that miR-134 was diversely expressed between normal and cancerous breast tissues. Besides, elevation of miR-134 could induce cell apoptosis by decreasing CREB and Bcl-2 in MCF7 breast cancer cells. To our knowledge, this is the first report that miR-134 had an effect on breast cancer.
Since miR-134 exerts a great role in embryonic stem cell differentiation and nervous system development [15-17], regulation of its expression was also an import part of its function exploration. We predicted and testified C/EBPα was an upstream promoting gene for miR-134. Then, as a target gene of miR-134, CREB was indirectly suppressed by C/EBPα. Thus the anti-apoptotic effect of CREB was inhibited and the cells exhibited proliferation inhibition. CREB acts as an anti-apoptotic gene in multiple tissues and the molecular mechanisms varies in different tissues [18-20]. However, we still do not know how CREB functions in the breast cancer cells. Further studies will be focused on the CREB mechanism of apoptosis protection in the breast cancer cells.
In conclusion, we found a novel pathway that C/EBPα induced breast cancer cell apoptosis via miR-134/CREB.
Acknowledgements
Heartfelt acknowledgements were uttered to the Xijing Hospital for their permission of using the specimens in this study.
Disclosure of conflict of interest
None.
References
- 1.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerød A Oslo Breast Cancer Consortium (OSBREAC) Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Børresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H, Yu Z, Liu S, Cui L, Chen X, Yao R. CUGBP1 promotes cell proliferation and suppresses apoptosis via down-regulating C/EBPα in human non-small cell lung cancers. Med Oncol. 2015;32:82. doi: 10.1007/s12032-015-0544-8. [DOI] [PubMed] [Google Scholar]
- 3.Chen BL, Sheu ML, Tsai KS, Lan KC, Guan SS, Wu CT, Chen LP, Hung KY, Huang JW, Chiang CK, Liu SH. CCAAT-Enhancer-Binding Protein Homologous Protein Deficiency Attenuates Oxidative Stress and Renal Ischemia-Reperfusion Injury. Antioxid Redox Signal. 2015;23:1233–45. doi: 10.1089/ars.2013.5768. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H, Imamura T, Fujiki A, Hirashima Y, Miyachi M, Inukai T, Hosoi H. Posttranscriptional modulation of C/EBPα prompts monocytic differentiation and apoptosis in acute myelomonocytic leukaemia cells. Leuk Res. 2012;36:735–41. doi: 10.1016/j.leukres.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhang ZC, Liu Y, Li SF, Guo L, Zhao Y, Qian SW, Wen B, Tang QQ, Li X. Suv39h1 mediates AP-2α-dependent inhibition of C/EBPα expression during adipogenesis. Mol Cell Biol. 2014;34:2330–8. doi: 10.1128/MCB.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloomer SA, Kregel KC, Brown KE. Heat stress stimulates hepcidin mRNA expression and C/EBPα protein expression in aged rodent liver. Arch Gerontol Geriatr. 2014;58:145–52. doi: 10.1016/j.archger.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gery S, Tanosaki S, Bose S, Bose N, Vadgama J, Koeffler HP. Down-regulation and growth inhibitory role of C/EBPα in breast cancer. Clin Cancer Res. 2005;11:3184–90. doi: 10.1158/1078-0432.CCR-04-2625. [DOI] [PubMed] [Google Scholar]
- 8.Bicker S, Khudayberdiev S, Weiß K, Zocher K, Baumeister S, Schratt G. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013;27:991–6. doi: 10.1101/gad.211243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr Opin Genet Dev. 2011;21:491–7. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Huang W, Cao J, Liu X, Meng F, Li M, Chen B, Zhang J. AMPK Plays a Dual Role in Regulation of CREB/BDNF Pathway in Mouse Primary Hippocampal Cells. J Mol Neurosci. 2015;56:782–8. doi: 10.1007/s12031-015-0500-2. [DOI] [PubMed] [Google Scholar]
- 12.Faraji F, Hu Y, Wu G, Goldberger NE, Walker RC, Zhang J, Hunter KW. An integrated systems genetics screen reveals the transcriptional structure of inherited predisposition to metastatic disease. Genome Res. 2014;24:227–40. doi: 10.1101/gr.166223.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-134/487b/655 Cluster Regulates TGF-β-Induced Epithelial-Mesenchymal Transition and Drug Resistance to Gefitinib by Targeting MAGI2 in Lung Adenocarcinoma Cells. Mol Cancer Ther. 2014;13:444–53. doi: 10.1158/1535-7163.MCT-13-0448. [DOI] [PubMed] [Google Scholar]
- 14.Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, Shapiro CL, Huebner K. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 2013;8:e55910. doi: 10.1371/journal.pone.0055910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garofalo M, Croce CM. Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev. 2015;81:53–61. doi: 10.1016/j.addr.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arámburo C, Alba-Betancourt C, Luna M, Harvey S. Expression and function of growth hormone in the nervous system: A brief review. Gen Comp Endocrinol. 2014;203:35–42. doi: 10.1016/j.ygcen.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 17.Meza-Sosa KF, Pedraza-Alva G, Pérez-Martínez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci. 2014;8:175. doi: 10.3389/fncel.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mylroie H, Dumont O, Bauer A, Thornton CC, Mackey J, Calay D, Hamdulay SS, Choo JR, Boyle JJ, Samarel AM, Randi AM, Evans PC, Mason JC. PKCϵ-CREB-Nrf2 signaling induces HO-1 in the vascular endothelium and enhances resistance to inflammation and apoptosis. Cardiovasc Res. 2015;106:509–19. doi: 10.1093/cvr/cvv131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou CH, Lai SL, Chen CN, Lee PH, Peng FC, Kuo ML, Lai HS. IL-6 regulates Mcl-1L expression through the JAK/PI3K/Akt/CREB signaling pathway in hepatocytes: implication of an anti-apoptotic role during liver regeneration. PLoS One. 2013;8:e66268. doi: 10.1371/journal.pone.0066268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Barbosa-Sampaio H, Jones PM, Persaud SJ, Muller DS. The CaMK4/CREB/IRS-2 cascade stimulates proliferation and inhibits apoptosis of β-cells. PLoS One. 2012;7:e45711. doi: 10.1371/journal.pone.0045711. [DOI] [PMC free article] [PubMed] [Google Scholar]