FIGURE 10.
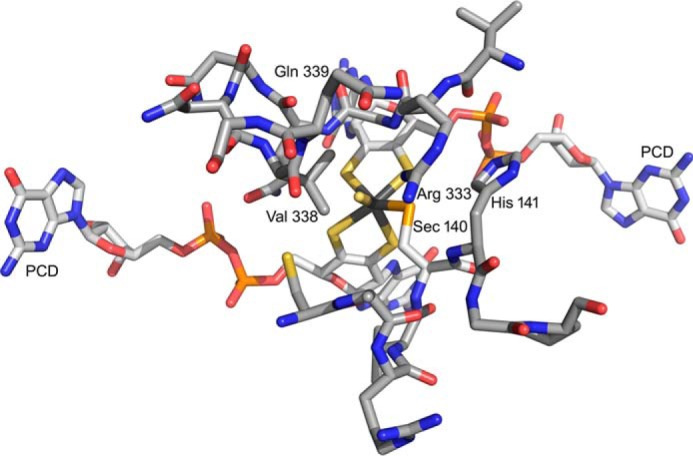
The active site of the FdhF formate dehydrogenase from E. coli (PDB code 1FDO). The molybdenum is at the center of the figure, coordinated by the two equivalents of pyranopterin cytidine dinucleotide (PCD), a terminal sulfido (represented as a coordinated water in the original PDB file), and Sec-140. The substrate binding site is delineated by two loops consisting of 137ARVUHGP143 and 328GVNPLRGQNNVQG340. Arg-333 has been proposed to interact with the negatively charged substrate. In these two stretches, only two residues differ in the FdsA subunit of the R. eutropha formate dehydrogenase: Sec-140, which is a Cys that coordinates the molybdenum in FdsA, and Val-338, which is a Leu in FdsA.
