FIGURE 2.
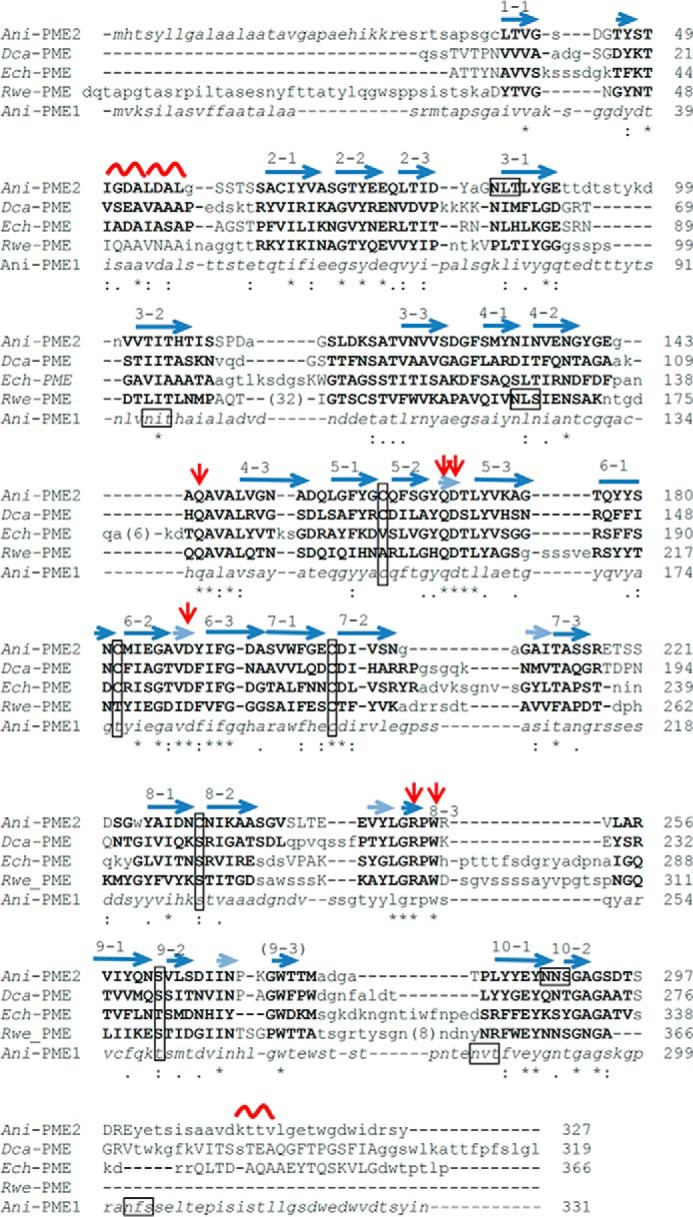
Adjusted ClustalW alignment of selected PME sequences. The raw sequence-based alignment has been adjusted to show structural alignment, where three-dimensional structural characterization has been made. Residues structurally homologous for fungal, plant, bacterial, and insect PMEs are highlighted in bold capitals; where structural homology is limited to two of the four structures light-face capitals are used. Residues in lowercase are not structurally homologous, and those in italics were not structurally characterized or observed. The secondary structure elements are noted above the pile-up of sequences. The β-helical turns are denoted n (n = 1–10); the strands comprising each turn are appended as n·1, n·2, and n·3; a series of very short β-strands, which form a 5-stranded β-sheet and encompass several active-site residues, are shown in a semi-transparent representation. The active-site Asp and Gln residues are flagged by red arrows; potential N-glycosylation motifs (NX(S/T)) of eukaryotic PMEs are boxed, and the ladder of cysteine and serine/threonine residues are boxed (vertical boxes). For clarity, extended insertions have been excised and the number of missing residues placed in parentheses.
