Fig. 2.
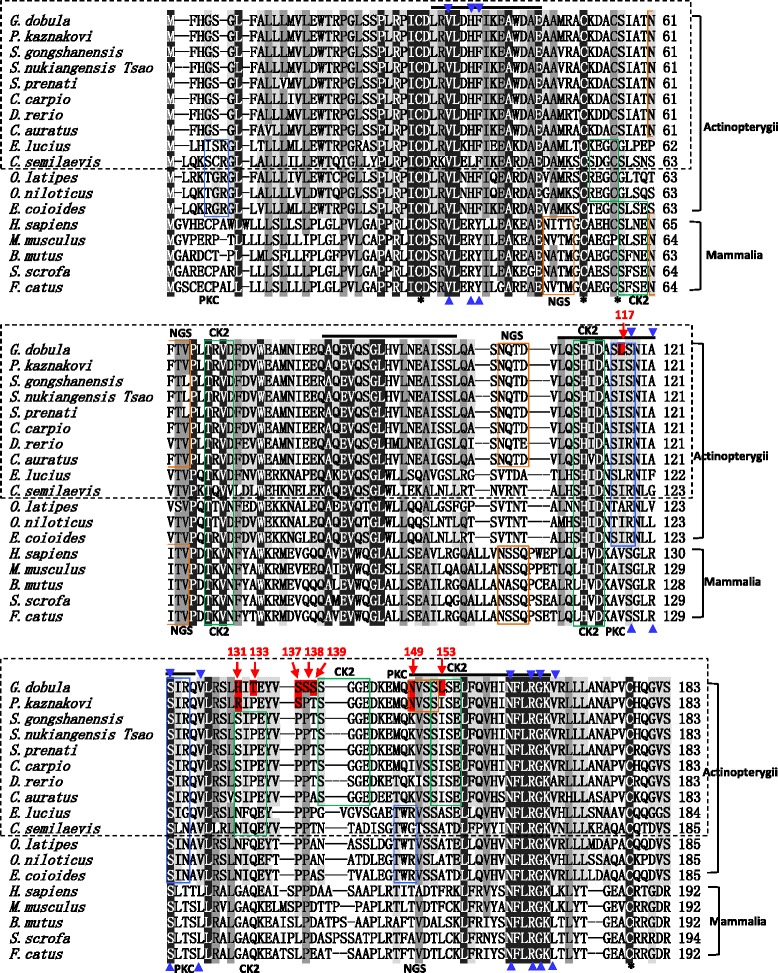
Sequence alignment of the EPOs. Seventeen EPO sequences, including those of 13 Actinopterygii fish species [G. dobula (KT188754), P. kaznakovi (KT188755), S. nukiangensis Tsao (KT188756), S. gongshanensis (KT188757), S. prenanti (KT188758), C. auratus (AGH20610), C. carpio (ABB83930), D. rerio (AAI62974), E. lucius (XP_010901758), C. semilaevis (XP_008325395), O. latipes (XP_004079700), O. niloticus (XP_003457688) and E. coioides (AAW29029)] and 5 mammals [S. scrofa (CAB96416), F. catus (AAA18282), B. mutus (XP_005910850), M. musculus (AAI44884) and H. sapiens (CAA26095)] were aligned with ClustalW. The ten species used for the phylogenetic tree construction (Fig. 3) are highlighted in a dashed rectangle. The limits of each helix are drawn according to the human EPO protein sequence and marked by bold lines. The four conserved cysteine residues are marked by asterisks. The NGS sites, the CK2 phosphorylation sites and the PKC phosphorylation sites are marked with orange, green and blue rectangles, respectively. The EPO receptor binding sites were inferred based on a structural model of human EPO [35] and are indicated by blue triangles. The deduced amino acid residues are numbered on the right. The amino acid sites unique to the two highly specialized schizothoracine fish G. dobula and P. kaznakovi within the Cyprinidae fishes are highlighted with red shadows and numbers
