Fig 1. Neurospora cells lacking the LSH/DDM1 homolog have normal DNA methylation but are hypersensitive to DNA damage.
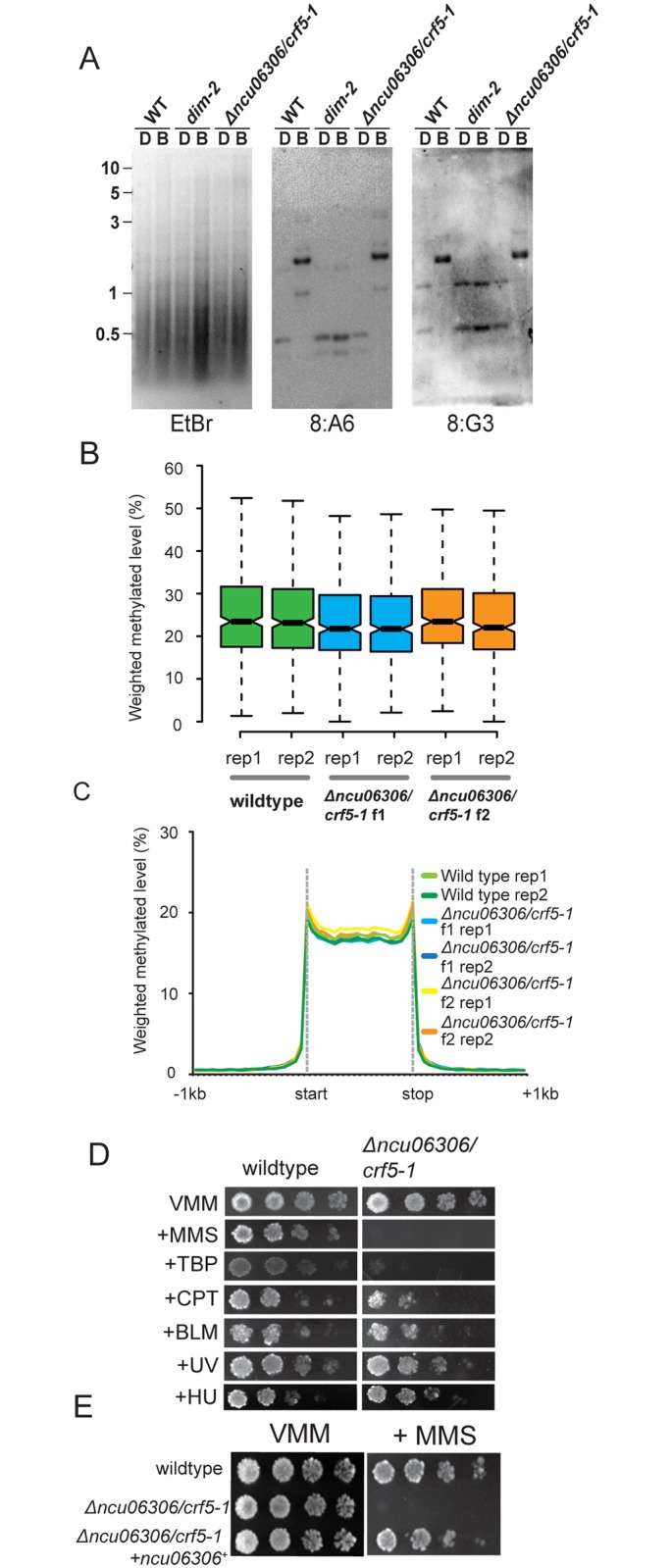
(A) Southern hybridizations with probes corresponding to the methylated 8:A6 and 8:G3 regions were performed using genomic DNA from the indicated strains digested with the cytosine-methylation-sensitive BfuCI [B] and–insensitive DpnII [D] restriction enzymes. The gel stained with Ethidium Bromide is also shown (EtBr). Numbers on the left correspond to the DNA ladder (kb). (B) Box plots showing the average methylation level for all methylated regions in wildtype, plus Δncu06306/crf5-1 strains from f1 and f2 generations. Two biological replicates are shown for each strain. The notches indicate the 95% confidence interval around the median. Overlapping notches indicate that the samples are not statistically different. (C) A metaplot showing the average distribution of DNA methylation across all methylated domains for wildtype and for Δncu06306/crf5-1 strains from f1 and f2 generations. (D) Serial dilutions of conidia (104−101) of wildtype and Δncu06306/crf5-1 were spotted on Vogel’s Minimal Medium (VMM) with or without the indicated genotoxic agents: methyl methanesulfonate (MMS; 0.025%), tert-Butyl hydroperoxide (TBP; 100 μM), camptothecin (CPT; 0.3 μg/mL), Bleomycin (BM; 0.2 μg/mL), Hydroxyurea (HU; 7mM). Cells were also exposed to Ultraviolet light (UV; 300 x 100mJ/cm2). (E) Introduction of wildtype ncu06306/crf5-1+ complements the MMS-hypersensitive phenotype of Δncu06306/crf5-1. Serial dilutions of the indicated strains were spotted on minimal medium (VMM) with or without 0.025% MMS.
