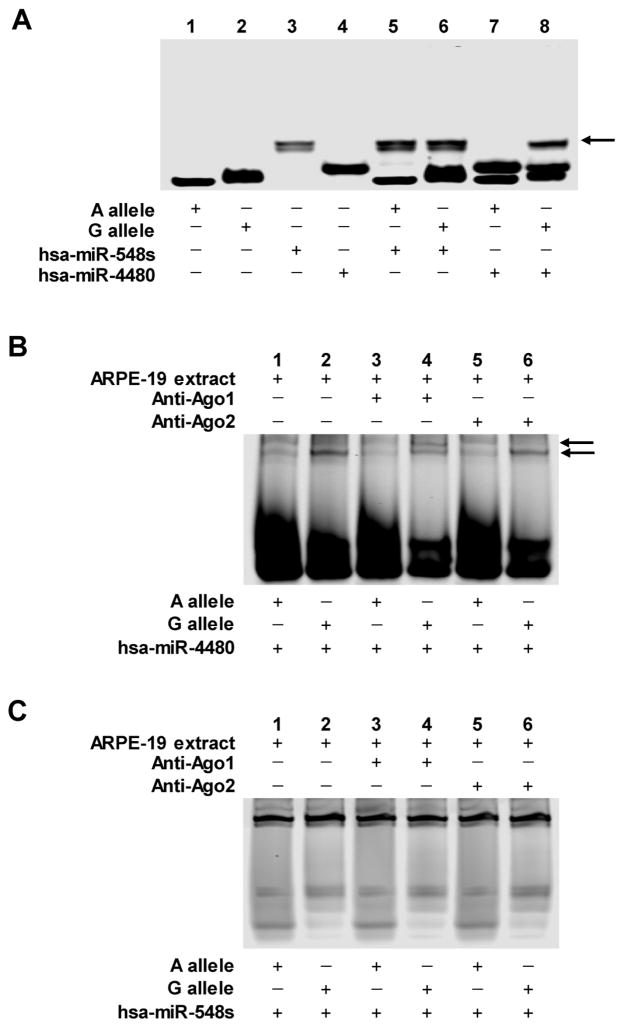
Figure 3.
Interaction of hsa-miR-548s and hsa-miR-4480 with IL-17A counterparts. (A) RNA EMSA was used to detect the formation of miRNA-mRNA complexes in vitro. Lanes 1–4 show the mobility of the labeled nucleotides. Lanes 5–6 show the mobility of hsa-miR-548s with each allele of rs7747909 in IL-17A. Lanes 7–8 show the mobility of hsa-miR-4480 with each allele of rs7747909 in IL-17A. (B) Addition of ARPE-19 cytoplasmic extracts to hsa-miR-4480 and IL-17A alleles shows formation of a RNA-protein complex (lanes 1–2). Ago1 (lanes 3–4) and Ago2 (lanes 5–6) antibodies were used to probe for Ago1 or Ago2 involvement in this RNA-protein complex via the formation of a supershift complex. (C) Addition of ARPE-19 cytoplasmic extracts to hsa-miR-548s and IL-17A alleles shows no formation of a RNA-protein complex (lanes 1–2). Ago1 (lanes 3–4) and Ago2 (lanes 5–6) antibodies were used to probe for Ago1 or Ago2 involvement in this RNA-protein complex via the formation of a supershift complex.