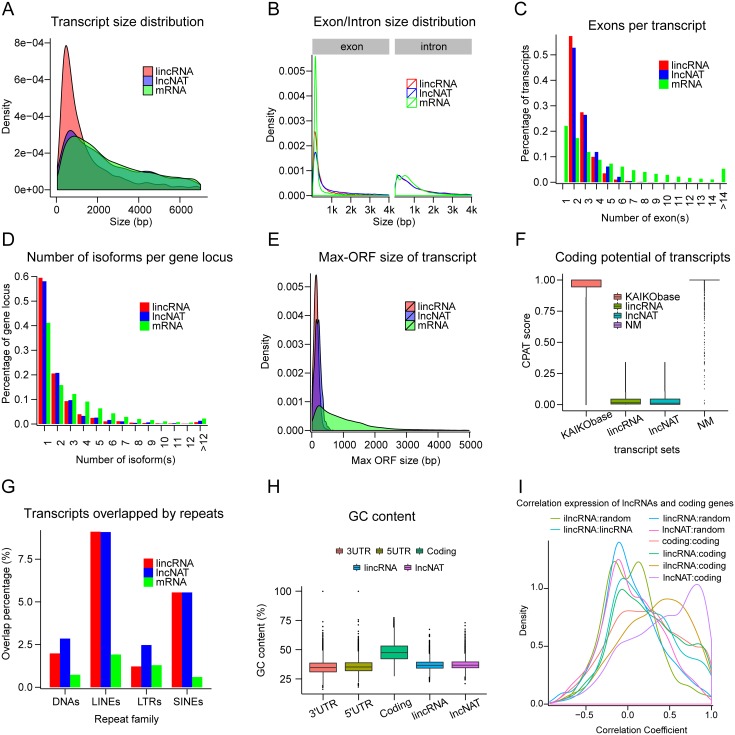
Fig 2. Features of silkworm lncRNAs.
(A) Transcript size distribution for lincRNAs, lncNATs, and mRNAs. (B) Exon (left) and intron (right) size distributions for lincRNA, lncNATs, and mRNAs (C) Number of exons per transcript for all lincRNAs, lncNATs, and mRNAs. (D) Distribution of the number of isoforms for each lincRNA, lncNAT, and mRNA gene locus. (E) Maximum ORF size distribution for lincRNAs, lncNATs, and mRNAs. (F) CPAT score distribution for KAIKObase gene model, ‘NM_’ reference sequence of silkworm, lincRNAs, and lncNATs. (G) Proportion of lincRNAs, lncNATs, and mRNAs transcripts covered by main repeat classes annotated by RepeatMasker. (H) GC content of lincRNAs, lncNATs, and mRNAs. (I) Pearson correlation coefficient distribution for neighboring transcript pairs from different datasets. Coding, protein-coding mRNAs; Random, random shuffle of mRNA positions.