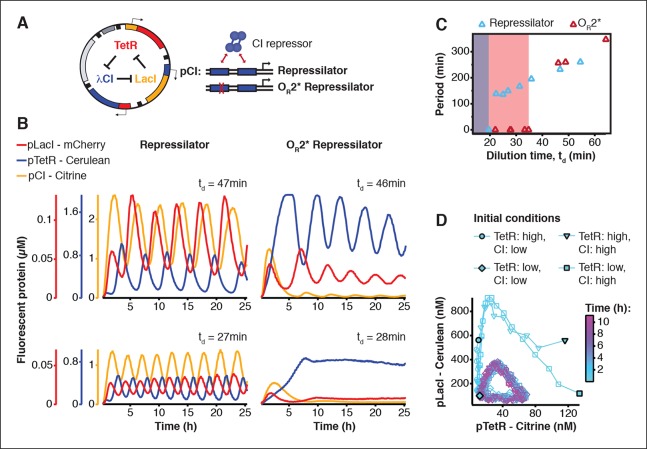
Figure 2. Cell-free repressilator characterization.
(A) Application of cell-free systems to characterize the original repressilator (Elowitz and Leibler, 2000) and a modified version with a point mutation in the CI promoter (OR2*) located in one of the binding sites of the CI repressor. (B) Expression from the three promoters of the repressilator and the OR2* version at different dilution times. (C) Oscillation periods of the repressilator as a function of dilution time. In the OR2* version sustained oscillations were supported in a narrower range of dilution times as compared to the original repressilator network. (D) Phase portrait of repressilator oscillations starting from different initial TetR and CI repressor concentrations.
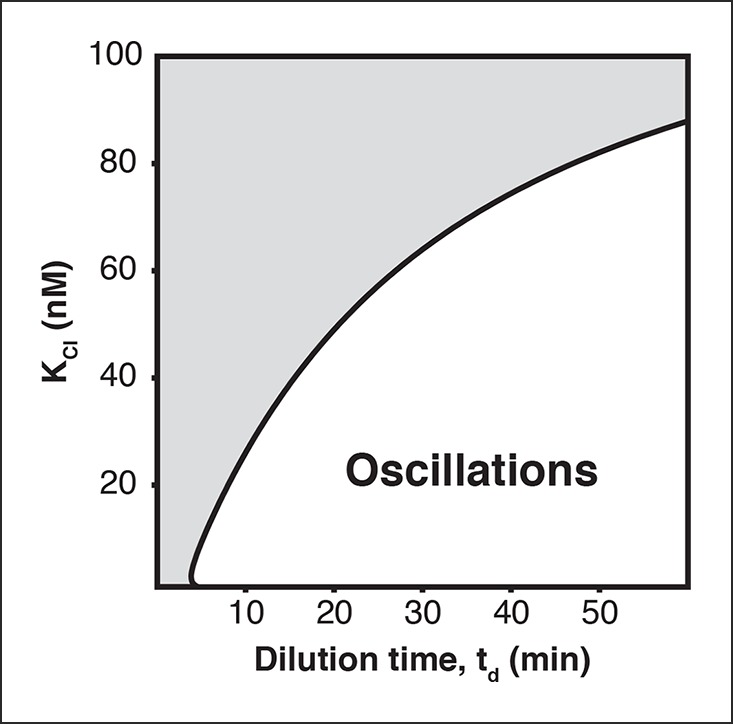
Figure 2—figure supplement 1. Oscillation parameter regime for a 3-node repressilator network in terms of dilution time and synthesis rates.
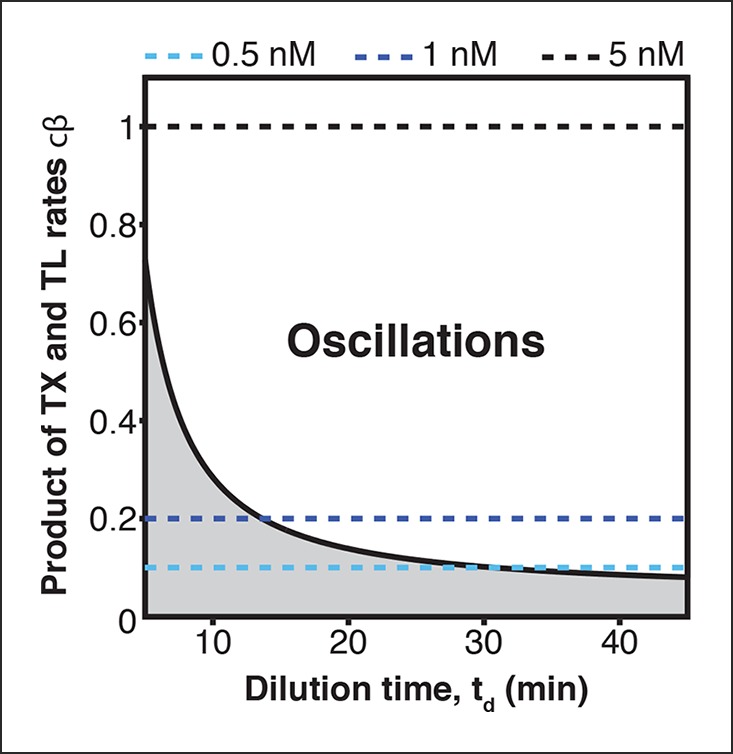
Figure 2—figure supplement 2. Existence of oscillations and periods for a 3-node repressilator network in terms of dilution time.
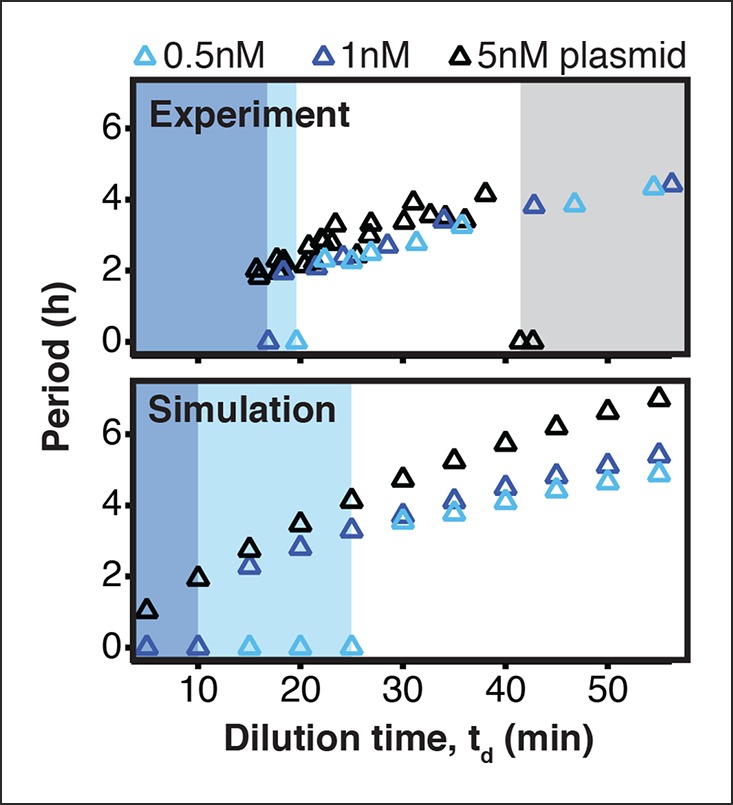
Figure 2—figure supplement 3. Oscillation parameter regime for a 3-node repressilator network in terms of dilution time and repressor binding affinity.