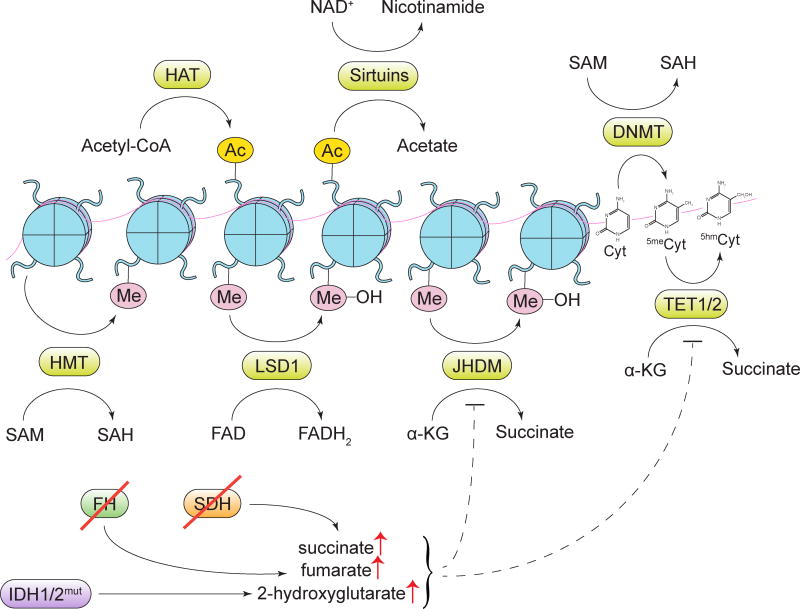
Figure 6. Alterations in metabolite-driven gene regulation.
Diverse metabolites serve as cofactors or substrates for enzymes involved in deposition and removal of epigenetic marks. HAT, histone acetyltransferase enzymes; Ac, an acetyl mark; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; DNMT, DNA methyltransferase enzymes; HMT, histone methyltransferase enzymes; Me, a methyl mark; LSD1, lysine-specific histone demethylase 1; JHDM, Jumonji domain-containing histone demethylase enzymes; Cyt, cytosine; 5meCyt, 5-methylcytosine; 5hmCyt, 5-hydroxymethylcytosine; TET1/2, ten-eleven translocation methylcytosine dioxygenase 1/2; α-KG, α-ketoglutarate; SDH, succinate dehydrogenase; FH, fumarate hydratase; IDH1/2, isocitrate dehydrogenase 1/2.