Fig. 3.
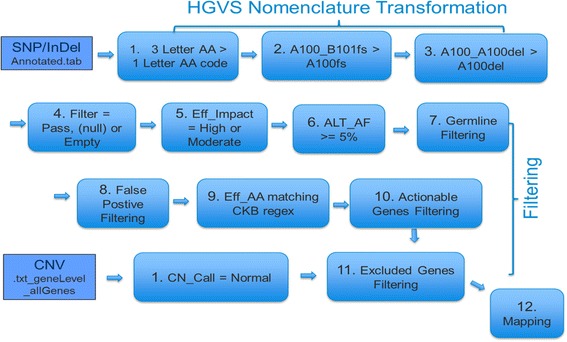
Automated variant mapping process. The genomic datasets are transformed to standard Human Genome Variation Society (HGVS) nomenclature, using a standardized Regular Expressions (regex) vocabulary incorporated into the JAX-CKB. This enables mapping and identification of clinically meaningful variants in a scalable and reproducible manner. The variant mapping program also includes several filtering steps, including removal of low-impact variants, those that did not pass metrics, and those that are likely germline or false-positive, to ensure reporting of only high-quality somatic variations
