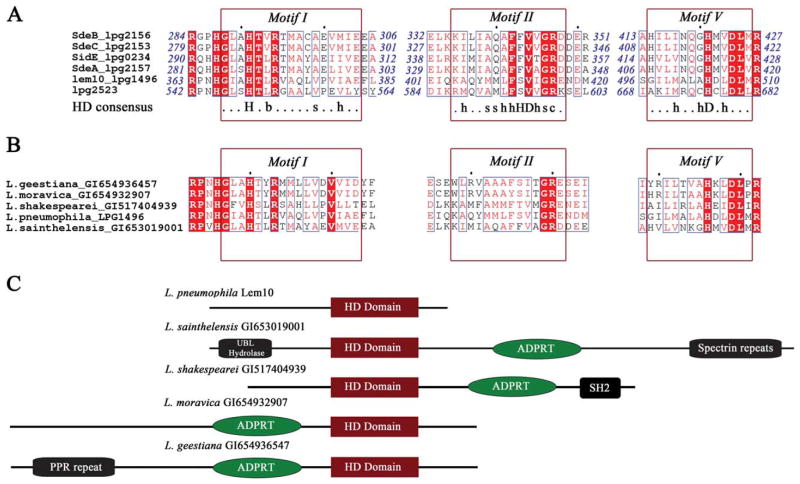
Figure 2.
A: Sequence alignment of Lem10 with homologous regions of other L. pneumophila effector proteins. Strictly conserved residues are highlighted in red, and functionally conserved residues are shown in red font. Sequence regions that correspond to the conserved motifs of the HD superfamily are indicated by red boxes, and the HD consensus sequence6 is shown under the alignment. In the consensus sequence, capital letters stand for residue names; lower case letters designate the following: b: bulky, s: small, h: hydrophobic, c: charged, x: any residue. B: Sequence alignment of Lem10 with homologous regions of proteins from other Legionella species. Annotation and color-coding as in (A). C: A cartoon representation of the predicted domain organization for the full sequences of the proteins containing homologous HD domains shown in (B).