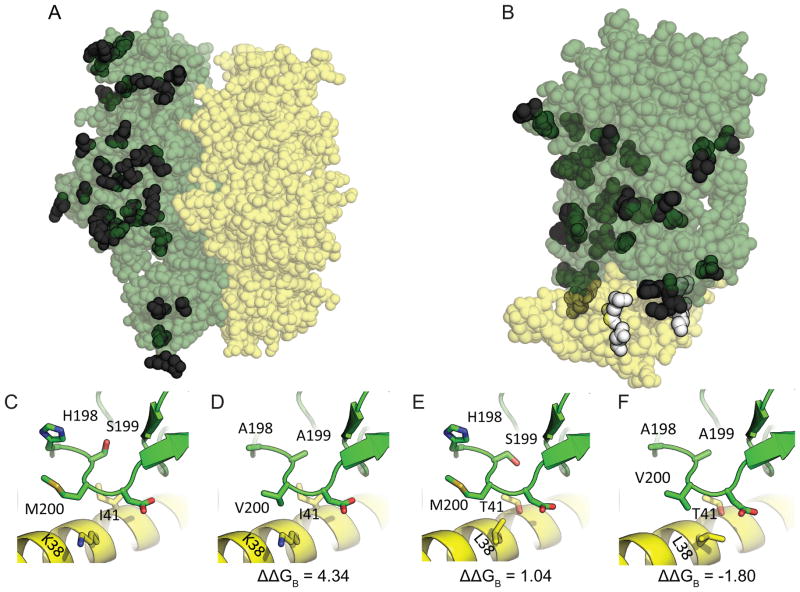
Figure 4.
Modeled structures for OXPHOS mitonuclear protein pairs based on ancestral Silene sequences for (A) ATP1-ATP2 and (B) COB-QCR7. (C) – (F) show residues in white indicated in (B), with (C) corresponding to the ancestral Silene sequences, (D) resulting from introducing the substitutions inferred to have taken place in the mitochondrial-encoded contact residues of S. conica, (E) resulting from introducing the substitutions inferred to have taken place in the nuclear-encoded contact residues of S. conica, and (F) resulting from introducing both mitochondrial and nuclear-encoded substitutions in the contact residues simultaneously. In (A) and (B) variable residues are opaque and conserved residues are transparent, while mitochondrial- and nuclear-encoded are green and yellow, respectively (or dark grey and light grey in the print version). In (C) – (F), changing variable contact residues are displayed as “sticks” while other residues are displayed as “ribbons”.